-Search query
-Search result
Showing 1 - 50 of 86 items for (author: zhang & yq)
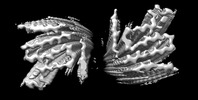
EMDB-36202: 
Cryo-EM structure of alpha-synuclein gS87 fibril

EMDB-36203: 
Cryo-EM structure of alpha-synuclein pS87 fibril

EMDB-34929: 
Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form

EMDB-34927: 
Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram

EMDB-35630: 
Cryo-EM structure of hMRS2-Mg

EMDB-35631: 
Cryo-EM structure of hMRS-highEDTA

EMDB-35632: 
Cryo-EM structure of hMRS2-lowEDTA

EMDB-35633: 
Cryo-EM structure of hMRS2-rest

EMDB-35522: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex(mask on receptor)

EMDB-35523: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex(mask on Giq-scFV16 complex)

EMDB-35524: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi1 complex(mask on receptor)

EMDB-35525: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi1 complex(mask on Gil-scFV16 complex)

EMDB-35529: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex (consensus map)

EMDB-35533: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex(consensus map)

EMDB-33884: 
Cryo-EM structure of Apo-alpha-syn fibril

EMDB-33890: 
Cryo-EM structure of dLAG3-alpha-syn fibril

EMDB-35356: 
Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex

EMDB-35357: 
Cryo-EM structure of the linoleic acid bound GPR120-Gi complex

EMDB-35358: 
Cryo-EM structure of the oleic acid bound GPR120-Gi complex

EMDB-35359: 
Cryo-EM structure of the TUG891 bound GPR120-Gi complex

EMDB-35360: 
Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex

EMDB-29736: 
Cryo-EM structure of the TUG891 bound GPR120-Giq complex

EMDB-27542: 
Cryo-EM reveals the molecular basis of laminin polymerization and LN-lamininopathies

EMDB-33236: 
The cryo-EM structure of Fe3+ induced alpha-syn fibril.

EMDB-33641: 
Cryo-EM structure of human sodium-chloride cotransporter

EMDB-33803: 
Cryo-EM structure of human sodium-chloride cotransporter

EMDB-33804: 
Cryo-EM structure of the C-terminal domain of the human sodium-chloride cotransporter

EMDB-33248: 
Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex

EMDB-33249: 
Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex

EMDB-33250: 
Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state

EMDB-31434: 
SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab

EMDB-30826: 
Cryo-EM structure of plant receptor like protein RXEG1 in complex with xyloglucanase XEG1 and BAK1

EMDB-32293: 
Cryo-EM structure of plant receptor like kinase NbBAK1 in RXEG1-BAK1-XEG1 complex

EMDB-32294: 
Plant receptor like protein RXEG1 in complex with xyloglucanase XEG1

EMDB-32295: 
Cryo-EM structure of plant receptor like protein RXEG1

EMDB-31435: 
SARS-CoV-2 S trimer in complex with 2 A5-10 Fabs (2 RBDs close and 1 RBD open)

EMDB-31436: 
SARS-CoV-2 S trimer in complex with 1 A5-10 Fab (3 RBDs close)

EMDB-31437: 
SARS-CoV-2 S trimer in complex with 3 A34-2 Fabs (3 RBDs open)

EMDB-31438: 
SARS-CoV-2 S trimer in complex with 2 A34-2 Fabs (2 RBDs open and 1 RBD close)

EMDB-31439: 
SARS-CoV-2 S trimer in complex with 3 A5-10 Fabs (2 RBDs close and 1 RBD open)
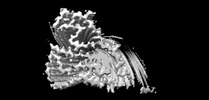
EMDB-33054: 
Cryo-EM structure of the TMEM106B fibril from normal elder

EMDB-33055: 
Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia

EMDB-31232: 
Structural basis for the tethered peptide activation of adhesion GPCRs

EMDB-31254: 
GPR114-Gs-scFv16 complex

EMDB-31033: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1)

EMDB-31209: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30

EMDB-31210: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 (local refinement of the RBD and Fab30)

EMDB-31035: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(state2, local refinement of the RBD and 35B5 Fab)

EMDB-31444: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab)

EMDB-31034: 
Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
