-Search query
-Search result
Showing 1 - 50 of 122 items for (author: walls & ac)
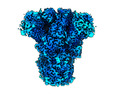
EMDB-26378: 
Cryo-EM structure of PDF-2180 Spike glycoprotein

EMDB-32686: 
Structure of NeoCOV RBD binding to Bat37 ACE2

EMDB-32693: 
Structure of PDF-2180-COV RBD binding to Bat37 ACE2

EMDB-27779: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit

PDB-8dya: 
Structure of the SARS-CoV-2 spike glycoprotein S2 subunit

EMDB-28558: 
SARS-CoV-2 BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment (local refinement of the RBD and S2X324)

EMDB-28559: 
SARS-CoV-2 Omicron BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment

PDB-8erq: 
SARS-CoV-2 BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment (local refinement of the RBD and S2X324)

PDB-8err: 
SARS-CoV-2 Omicron BA.1 spike ectodomain trimer in complex with the S2X324 neutralizing antibody Fab fragment
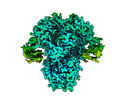
EMDB-26727: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation
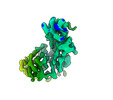
EMDB-26729: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)
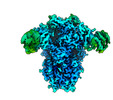
EMDB-26730: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
EMDB-26731: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)

PDB-7us6: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation

PDB-7us9: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)

PDB-7usa: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation

PDB-7usb: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)

EMDB-24678: 
SARS-CoV-2 S/S2M11/SNAP1 Global Refinement

EMDB-24679: 
SARS-CoV-2 S/S2M11/SNAP1 Local Refinement

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175

EMDB-25990: 
SARS-CoV-2 S B.1.1.529 Omicron variant (RBD + S309 Local Refinement)

EMDB-25991: 
SARS-CoV-2 S NTD B.1.1.529 Omicron variant + S309 Local Refinement

EMDB-25992: 
SARS-CoV-2 S B.1.1.529 Omicron variant + S309 + S2L20 Global Refinement

EMDB-25993: 
SARS-CoV-2 S B.1.1.529 Omicron variant + S309 + S2L20 Global Refinement (Two-open RBDs and one-closed RBD)

EMDB-25785: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (three receptor-binding domains open)

EMDB-25783: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (local refinement of the RBD and S2K146)

EMDB-25784: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (two receptor-binding domains open)

PDB-7tas: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (local refinement of the RBD and S2K146)

PDB-7tat: 
SARS-CoV-2 spike in complex with the S2K146 neutralizing antibody Fab fragment (two receptor-binding domains open)

EMDB-25263: 
SARS-CoV-2 S B.1.617.2 delta variant + S2M11 + S2L20 Global Refinement

EMDB-25264: 
SARS-CoV-2 S NTD B.1.617.2 delta variant + S2L20 Local Refinement

EMDB-25265: 
SARS-CoV-2 S B.1.617.1 kappa variant + S309 + S2L20 Global Refinement

EMDB-25266: 
SARS-CoV-2 S RBD B.1.617.1 kappa variant S309 Local Refinement

EMDB-25267: 
SARS-CoV-2 S NTD B.1.617.1 kappa variant S2L20 Local Refinement

EMDB-25268: 
SARS-CoV-2 S B.1.617.1 kappa variant + S2X303 Global Refinement

EMDB-25269: 
SARS-CoV-2 S NTD B.1.617.1 kappa variant S2X303 Local Refinement

EMDB-24607: 
SARS-CoV-2 spike in complex with the S2X58 neutralizing antibody Fab fragment (two receptor-binding domains open)

EMDB-24608: 
SARS-CoV-2 spike in complex with the S2X58 neutralizing antibody Fab fragment (three receptor-binding domains open)

EMDB-24533: 
SARS-CoV-2 spike protein bound to the S2P6 and S2M11 Fab fragments

EMDB-24347: 
SARS-CoV-2 S glycoprotein in complex with S2X259 Fab

EMDB-24365: 
SARS-CoV-2 S bound to S2X259 Fab (local refinement of the RBD/S2X259 variable domains)

PDB-7ra8: 
SARS-CoV-2 S glycoprotein in complex with S2X259 Fab

PDB-7ral: 
SARS-CoV-2 S bound to S2X259 Fab (local refinement of the RBD/S2X259 variable domains)

EMDB-24299: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment (local refinement of the RBD and S2D106)

EMDB-24300: 
SARS-CoV-2 spike in complex with the S2D106 neutralizing antibody Fab fragment
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
