-Search query
-Search result
Showing all 46 items for (author: shen & yp)

EMDB-36960: 
F8-A22-E4 complex of MPXV in complex with DNA and Ara-CTP

EMDB-36962: 
F8-A22-E4 complex of MPXV in complex with DNA and dCTP

EMDB-36963: 
the local map of DNA and Ara-CTP binding site

EMDB-36964: 
the local map of DNA and dCTP binding site
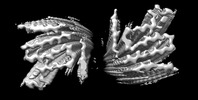
EMDB-36202: 
Cryo-EM structure of alpha-synuclein gS87 fibril

EMDB-36203: 
Cryo-EM structure of alpha-synuclein pS87 fibril

EMDB-35264: 
S-ECD (Omicron BA.2.75) in complex with PD of ACE2

EMDB-35266: 
S-ECD (Omicron BF.7) in complex with PD of ACE2

EMDB-35267: 
S-ECD (Omicron XBB.1) in complex with PD of ACE2

EMDB-35268: 
S-RBD(Omicron BA.3) in complex with PD of ACE2

EMDB-35269: 
S-RBD (Omicron BA.2.75) in complex with PD of ACE2

EMDB-35270: 
S-RBD (Omicron BF.7) in complex with PD of ACE2

EMDB-35272: 
S-RBD (Omicron XBB.1) in complex with PD of ACE2

EMDB-35254: 
ACE2-SIT1 complex bound with proline

EMDB-35255: 
ACE2-B0AT1 complex bound with glutamine

EMDB-35256: 
ACE2-B0AT1 complex bound with methionine

EMDB-35260: 
Cryo-EM map of the ACE2-SIT1 complex bound with proline, focused refined on extracellular region

EMDB-35261: 
cryo-EM map of the ACE2-SIT1 complex bound with proline, focused refined on transmembrane region

EMDB-35262: 
cryo-EM map of the ACE2-B0AT1 complex bound with glutamine, focused refined on extracellular region

EMDB-35265: 
cryo-EM map of the ACE2-B0AT1 complex bound with glutamine, focused refined on transmembrane region

EMDB-35271: 
cryo-EM map of the ACE2-B0AT1 complex bound with methionine, focused refined on extracellular region

EMDB-35273: 
cryo-EM map of the ACE2-B0AT1 complex bound with methionine, focused refined on transmembrane region

EMDB-33577: 
S-ECD (Omicron BA.3) in complex with two PD of ACE2

EMDB-33578: 
S-ECD (Omicron BA.5) in complex with PD of ACE2

EMDB-33575: 
S-ECD (Omicron BA.2) in complex with PD of ACE2

EMDB-33576: 
S-ECD (Omicron BA.3) in complex with three PD of ACE2

EMDB-33579: 
map focused on the interface between BA.2 RBD and ACE2

EMDB-33580: 
map focused on the interface between BA.3 RBD and ACE2
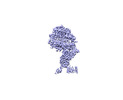
EMDB-33581: 
map focused on the interface between BA.4 RBD and ACE2

EMDB-34885: 
F8-A22-E4 complex in hexameric form local map

EMDB-34886: 
F8-A22-E4 complex of MPXV in hexameric form

EMDB-34887: 
F8-A22-E4 complex of MPXV in trimeric form

EMDB-33653: 
SIT1-ACE2-BA.5 RBD

EMDB-33656: 
Local map of the interface of ACE2 and BA.5 RBD

EMDB-33657: 
TM domain of SIT1

EMDB-33652: 
SIT1-ACE2-BA.2 RBD

EMDB-33654: 
Local map of ACE2 and BA.2 interface
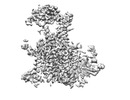
EMDB-33655: 
TM domain of SIT1
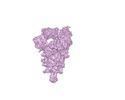
EMDB-33202: 
S-ECD (Omicron) in complex with STS165
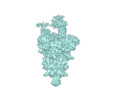
EMDB-33203: 
S-ECD (Omicron) in complex with PD of ACE2

EMDB-33204: 
S-ECD (Omicron) in complex with PD of ACE2 focused on RBD_PD sub-complex
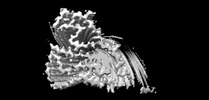
EMDB-33054: 
Cryo-EM structure of the TMEM106B fibril from normal elder

EMDB-33055: 
Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia

EMDB-31249: 
S protein of SARS-CoV-2 in complex with GW01

EMDB-31250: 
Local map of S protein of SARS-CoV-2 in complex with GW01 Focused on RND-GW01 sub_complex

EMDB-32092: 
Cryo-EM structure of amyloid fibril formed by FUS low complexity domain
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
