-Search query
-Search result
Showing 1 - 50 of 37,855 items for (database: EMDB)

EMDB-41903: 
Cryo-EM structure of PsBphP in Pr state

EMDB-41941: 
Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers FL

EMDB-41942: 
Cryo-EM structure of PsBphP in Pfr state, Dimer of Dimers PSM only

EMDB-41943: 
Cryo-EM structure of PsBphP in Pfr state, medial PSM only

EMDB-41944: 
Cryo-EM structure of PsBphP in Pfr state, splayed PSM only

EMDB-42030: 
Cryo-EM structure of PsBphP in Pr state, extended DHp

EMDB-44372: 
In-cell Saccharomyces cerevisiae nuclear pore complex with single nuclear ring

EMDB-44377: 
In-cell Saccharomyces cerevisiae nuclear pore complex with double nuclear ring and basket

EMDB-44379: 
In-cell Mus musculus nuclear pore complex with nuclear basket

EMDB-44381: 
In-cell Toxoplasma gondii nuclear pore complex

EMDB-45197: 
In-cell Saccharomyces cerevisiae symmetry-expanded nuclear pore complex with double nuclear ring and basket

EMDB-45198: 
In-cell Saccharomyces cerevisiae symmetry-expanded nuclear pore complex with single nuclear ring

EMDB-45199: 
In-cell Saccharomyces cerevisiae nuclear pore complex cytoplasmic ring focused refinement

EMDB-45200: 
In-cell Saccharomyces cerevisiae nuclear pore complex inner ring focused refinement

EMDB-45201: 
In-cell Saccharomyces cerevisiae nuclear pore complex single nuclear ring focused refinement

EMDB-45202: 
In-cell Saccharomyces cerevisiae nuclear pore complex double nuclear ring focused refinement

EMDB-45203: 
In-cell Saccharomyces cerevisiae nuclear pore complex nuclear basket focused refinement

EMDB-45204: 
In-cell Saccharomyces cerevisiae nuclear pore complex membrane focused refinement for single nuclear ring

EMDB-45205: 
In-cell Saccharomyces cerevisiae nuclear pore complex membrane focused refinement for double nuclear ring

EMDB-45216: 
In-cell Mus musculus nuclear pore complex with nuclear basket consensus map

EMDB-45219: 
In-cell Mus musculus nuclear pore complex cytoplasmic ring focused refinement

EMDB-45220: 
In-cell Mus musculus nuclear pore complex inner ring focused refinement

EMDB-45222: 
In-cell Mus musculus nuclear pore complex nuclear ring focused refinement

EMDB-45223: 
In-cell Mus musculus nuclear pore complex basket focused refinement

EMDB-45227: 
In-cell Mus musculus nuclear pore complex membrane focused refinement

EMDB-45228: 
In-cell Toxoplasma gondii symmetry-expanded nuclear pore complex
EMDB-45255: 
In-cell Saccharomyces cerevisiae C8-symmetrised nuclear pore complex consensus map

EMDB-45256: 
In-cell Saccharomyces cerevisiae symmetry-expanded nuclear pore complex consensus map

EMDB-45257: 
In-cell Mus musculus nuclear pore complex with nuclear basket consensus map

EMDB-45258: 
In-cell Mus musculus symmetry-expanded nuclear pore complex with nuclear basket consensus map

EMDB-45259: 
In-cell Toxoplasma gondii C8-symmetrised nuclear pore complex consensus map
EMDB-42603: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/hexamer)
EMDB-42625: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC804515)
EMDB-42626: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/up)
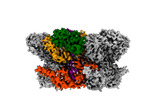
EMDB-42627: 
Human p97/VCP R155H mutant structure with a triazole inhibitor (NSC819701/down)
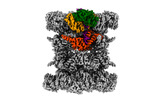
EMDB-44748: 
Human p97/VCP structure with a triazole inhibitor (NSC799462/dodecamer)
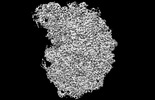
EMDB-45185: 
Cutibacterium acnes 50S ribosomal subunit with Clindamycin bound

EMDB-43514: 
SPOT-RASTR - a cryo-EM specimen preparation technique that overcomes problems with preferred orientation and the air/water interface
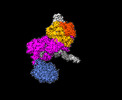
EMDB-42842: 
A Mitochondrial Replication Complex: PolG/PolG2 Bound to DNA in complex with the Single-Stranded Binding Protein (mtSSB)

EMDB-41766: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant, scFv16, and dopamine

EMDB-41776: 
CryoEM structure of D2 dopamine receptor in complex with GoA KE mutant and dopamine

EMDB-44551: 
Map of eastern equine encephalitis virus q3 spike protein in complex with VLDLR without masked refinement
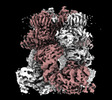
EMDB-44255: 
Cryo-EM structure of the mouse TRPM8 channel in the ligand-free desensitized state

EMDB-44256: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014

EMDB-44257: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMG2850
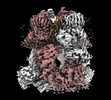
EMDB-44258: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist AMTB
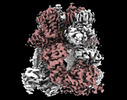
EMDB-44259: 
Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3
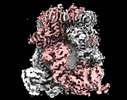
EMDB-44260: 
Cryo-EM structure of the avian great tit TRPM8 channel in complex with the antagonist TC-I 2014

EMDB-44261: 
Cryo-EM structure of the mouse TRPM8 channel in complex with PI(4,5)P2 and Ca2+
EMDB-44262: 
Cryo-EM structure of the mouse TRPM8 channel in complex with Ca2+ in the absence of PI(4,5)P2
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
