-Search query
-Search result
Showing 1 - 50 of 2,276 items for (author: zheng & y)

EMDB-37362: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

EMDB-37363: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

PDB-8w9a: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W

PDB-8w9b: 
CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site
Method: single particle / : Huang X, Ren X, Zhong W
EMDB-36808: 
Cryo-EM structure of KEOPS complex from Arabidopsis thaliana
Method: single particle / : Zheng XX, Zhu L, Duan L, Zhang WH

PDB-8k20: 
Cryo-EM structure of KEOPS complex from Arabidopsis thaliana
Method: single particle / : Zheng XX, Zhu L, Duan L, Zhang WH
EMDB-35732: 
Cryo-EM structure of SARS-CoV-2 spike protein in complex with 1C4
Method: single particle / : Sun H, Jiang Y, Zheng Q, Li S, Xia N

EMDB-36712: 
Histamine-bound H4R/Gi complex
Method: single particle / : He Y, Xia R

EMDB-36714: 
Clozapine-bound H4R/Gi complex
Method: single particle / : He Y, Xia R

EMDB-36715: 
VUF6884-bound H4R/Gi complex
Method: single particle / : He Y, Xia R

EMDB-36716: 
Clobenpropit-bound H4R/Gi complex
Method: single particle / : He Y, Xia R

EMDB-35832: 
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Method: single particle / : Jia GW, Wang X, Zhang CB, Dong HH, Su ZM

PDB-8iyx: 
Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365
Method: single particle / : Jia GW, Wang X, Zhang CB, Dong HH, Su ZM

EMDB-41071: 
Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B
Method: single particle / : Tonggu L, Wisedchaisri G, Gamal El-Din TM, Zheng N, Catterall WA

PDB-8t6l: 
Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B
Method: single particle / : Tonggu L, Wisedchaisri G, Gamal El-Din TM, Zheng N, Catterall WA

EMDB-37516: 
BA.2(S375) Spike (S6P)/hACE2 complex
Method: single particle / : Wei X, Zhang Z

EMDB-37517: 
Local refinement of RBD-ACE2
Method: single particle / : Wei X, Zhang Z

EMDB-36369: 
Focused map on the aCTD-AfsR(T337A) region of the Streptomyces coelicolor RNAP-promoter open complex with AfsR(T337A) dimer
Method: single particle / : Wang Y, Zheng J

EMDB-36370: 
AfsR(T337A) transcription activation complex
Method: single particle / : Wang Y, Zheng J
EMDB-37881: 
GPR3/Gs complex
Method: single particle / : He Y, Xiong Y

EMDB-33990: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N
EMDB-33992: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N
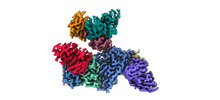
EMDB-33993: 
Cryo-EM density map of EBV gHgL-gp42 in complex with four mAbs 5E3, 3E8, 6H2 and 10E4
Method: single particle / : Liu L, Sun H, Jiang Y, Liu X, Zhao D, Zheng Q, Li S, Chen Y, Xia N
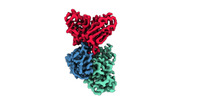
EMDB-33994: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

PDB-7yoy: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

PDB-7yp1: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

PDB-7yp2: 
Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement)
Method: single particle / : Liu L, Sun H, Jiang Y, Hong J, Zheng Q, Li S, Chen Y, Xia N

EMDB-34928: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 S-trimer in complex with fab L4.65 and L5.34
Method: single particle / : Gao GF, Liu S

EMDB-34940: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.4 S-trimer in complex with fab L4.65 and L5.34
Method: single particle / : Gao GF, Liu S

EMDB-34945: 
Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34
Method: single particle / : Gao GF, Liu S

PDB-8hp9: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2 S-trimer in complex with fab L4.65 and L5.34
Method: single particle / : Gao GF, Liu S

PDB-8hpq: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.4 S-trimer in complex with fab L4.65 and L5.34
Method: single particle / : Gao GF, Liu S

PDB-8hpv: 
Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34
Method: single particle / : Gao GF, Liu S

EMDB-35688: 
Cas6f of Csy-AcrIF4-dsDNA
Method: single particle / : Feng Y, Zhang LX

EMDB-35689: 
Cas8f of Csy-AcrIF4-dsDNA
Method: single particle / : Feng Y, Zhang LX

EMDB-35690: 
Overall structure of Csy-AcrIF4-dsDNA
Method: single particle / : Feng Y, Zhang LX

EMDB-29794: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29795: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29796: 
SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3
Method: single particle / : Ye G, Bu F, Liu B, Li F

EMDB-29797: 
SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g72: 
SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A)
Method: single particle / : Ye G, Bu F, Liu B, Li F

PDB-8g73: 
SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A
Method: single particle / : Ye G, Bu F, Liu B, Li F
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search











 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
