-Search query
-Search result
Showing 1 - 50 of 91 items for (author: zalk & r)

EMDB-17369: 
A CHIMERA construct containing human SARM1 ARM and SAM domains and C. elegans TIR domain.
Method: single particle / : Isupov MN, Opatowsky Y

EMDB-17370: 
C. elegans TIR-1 protein.
Method: single particle / : Isupov MN, Opatowsky Y

PDB-8p2l: 
A CHIMERA construct containing human SARM1 ARM and SAM domains and C. elegans TIR domain.
Method: single particle / : Isupov MN, Opatowsky Y

PDB-8p2m: 
C. elegans TIR-1 protein.
Method: single particle / : Isupov MN, Opatowsky Y

EMDB-15447: 
Cryo-electron tomogram of Butyrivibrio fibrisolvens CF3
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-16477: 
Cryo-electron tomogram of an intact Bacteroides cellulosyliticus CRE21 cell (DSM 14838)
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-15445: 
Cryo-electron tomogram of Agathobacter ruminis DSM 29029
Method: electron tomography / : Wimmer BH

EMDB-15448: 
Cryo-electron tomogram of Pseudobutyrivibrio sp. LB2011
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-15449: 
Cryo-electron tomogram of Wolinella sp. ATCC 33567
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-16474: 
Cryo-electron tomogram of an intact Bacteroides caccae DSM 19024 cell.
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-16478: 
Cryo-electron tomogram of an intact Lachnospira multipara G6 (ATCC 19207) cell.
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-16479: 
Cryo-electron tomogram of a Roseburia intestinalis DSM 14610 cell.
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-16488: 
Cryo-electron tomogram of an intact Bacteroides thetaiotaomicron DSM 2079 cell.
Method: electron tomography / : Wimmer BH, Medalia O

EMDB-13951: 
Inhibitor-induced hSARM1 duplex
Method: single particle / : Zalk R, Kahzma T, Guez-Haddad J

PDB-7qg0: 
Inhibitor-induced hSARM1 duplex
Method: single particle / : Zalk R, Kahzma T, Guez-Haddad J

EMDB-12809: 
MAT in complex with SAMH
Method: single particle / : Simon H, Kleiner D, Shmulevich F, Zarivach R, Zalk R, Tang H, Ding F, Bershtein S

PDB-7ock: 
MAT in complex with SAMH
Method: single particle / : Simon H, Kleiner D, Shmulevich F, Zarivach R, Zalk R, Tang H, Ding F, Bershtein S

EMDB-11365: 
M6A and sediment structure inside iron-filled ferritin-M6A without symmetry assumption
Method: single particle / : Davidov G, Abelya G, Zalk R, Zarivach R, Frank GA

EMDB-11272: 
Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Meyron Holtz EG, Zarivach R, Frank GA

EMDB-11304: 
radiation damage-assisted analysis of iron-filled ferritin-M6A.
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Shagidov D, Meyron Holtz EG, Zarivach R, Frank GA

PDB-6zlq: 
Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Meyron Holtz EG, Zarivach R, Frank GA

EMDB-11265: 
Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Meyron Holtz EG, Zarivach R, Frank GA

PDB-6zlg: 
Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Meyron Holtz EG, Zarivach R, Frank GA

EMDB-12761: 
Anthrax toxin prepore in complex with the neutralizing Fab cAb29
Method: single particle / : Hoelzgen F, Zalk R, Alcalay R, Cohen-Schwartz S, Garau G, Shahar A, Mazor O, Frank GA

PDB-7o85: 
Anthrax toxin prepore in complex with the neutralizing Fab cAb29
Method: single particle / : Hoelzgen F, Zalk R, Alcalay R, Cohen-Schwartz S, Garau G, Shahar A, Mazor O, Frank GA

EMDB-11214: 
Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Meyron Holtz EG, Zarivach R, Frank GA

PDB-6zh5: 
Folding of an iron binding peptide in response to sedimentation is resolved using ferritin as a nano-reactor
Method: single particle / : Davidov G, Abelya G, Zalk R, Izbicki B, Shaibi S, Spektor L, Meyron Holtz EG, Zarivach R, Frank GA

EMDB-10625: 
Cin8 motor domain (short loop 8) bound to microtubules
Method: helical / : Siegler N, Singh SK, Zalk R, Debs GE, Sindelar CV, Gheber L, Zarivach R

EMDB-11187: 
hSARM1 GraFix-ed
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Mim C, Isupov MN, Zalk R, Hons M, Opatowsky Y

PDB-6zfx: 
hSARM1 GraFix-ed
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Mim C, Isupov MN, Zalk R, Hons M, Opatowsky Y
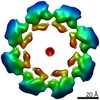
EMDB-11190: 
SARM1 SAM1-2 domains
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Mim C, Isupov MN, Zalk R, Hons M, Opatowsky Y

EMDB-11191: 
SARM1 SAM1-2 domains
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Mim C, Isupov MN, Zalk R, Hons M, Opatowsky Y

EMDB-11834: 
hSARM1 NAD+ complex
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Mim C, Isupov MN, Zalk R, Dessau M, Hons M, Opatowsky Y

PDB-6zg0: 
SARM1 SAM1-2 domains
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Mim C, Isupov MN, Zalk R, Hons M, Opatowsky Y

PDB-6zg1: 
SARM1 SAM1-2 domains
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Dessau M, Mim C, Isupov MN, Zalk R, Hons M, Opatowsky Y

PDB-7anw: 
hSARM1 NAD+ complex
Method: single particle / : Sporny M, Guez-Haddad J, Khazma T, Yaron A, Mim C, Isupov MN, Zalk R, Dessau M, Hons M, Opatowsky Y

EMDB-4648: 
Packaging of DNA Origami in Viral Capsids
Method: single particle / : Kopatz I, Zalk R, Levi-Kalisman Y, Zlotkin-Rivkin E, Frank GA, Kler S

EMDB-4651: 
Packaging of DNA Origami in Viral Capsids
Method: single particle / : Kopatz I, Zalk R, Levi-Kalisman Y, Zlotkin-Rivkin E, Frank GA, Kler S
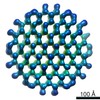
EMDB-4652: 
Packaging of DNA Origami in Viral Capsids
Method: single particle / : Kopatz I, Zalk R, Levi-Kalisman Y, Zlotkin-Rivkin E, Frank GA, Kler S

EMDB-4653: 
Packaging of DNA Origami in Viral Capsids
Method: single particle / : Kopatz I, Zalk R, Levi-Kalisman Y, Zlotkin-Rivkin E, Frank GA, Kler S

EMDB-8342: 
Structural basis for gating and activation of RyR1 (30 uM Ca2+ dataset, all particles)
Method: single particle / : Clarke OB, des Georges A

EMDB-8372: 
Structure of rabbit RyR1 (Ca2+-only dataset, class 1)
Method: single particle / : Clarke OB, des Georges A

EMDB-8373: 
Structure of rabbit RyR1 (Ca2+-only dataset, class 2)
Method: single particle / : Clarke OB, des Georges A

EMDB-8374: 
Structure of rabbit RyR1 (Ca2+-only dataset, class 3)
Method: single particle / : Clarke OB, des Georges A

EMDB-8375: 
Structure of rabbit RyR1 (Ca2+-only dataset, class 4)
Method: single particle / : Clarke OB, des Georges A, Zalk R, Marks AR, Hendrickson WA, Frank J

EMDB-8376: 
Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1)
Method: single particle / : Clarke OB, des Georges A, Zalk R, Marks AR, Hendrickson WA, Frank J

EMDB-8377: 
Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 2)
Method: single particle / : Clarke OB, des Georges A, Zalk R, Marks AR, Hendrickson WA, Frank J

EMDB-8378: 
Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1&2)
Method: single particle / : Clarke OB, des Georges A

EMDB-8379: 
Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 4)
Method: single particle / : Clarke OB, des Georges A, Zalk R, Marks AR, Hendrickson WA, Frank J

EMDB-8380: 
Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 3)
Method: single particle / : Clarke OB, des Georges A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
