-Search query
-Search result
Showing 1 - 50 of 896 items for (author: zak & o)

EMDB-18482: 
Herpes simplex virus 1 capsid (WT) vertices in perinuclear NEC-coated vesicles determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18484: 
Herpes simplex virus 1 nuclear egress complex (WT) determined in situ from perinuclear vesicles
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-17974: 
Pseudorabies virus cytosolic C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-17975: 
Pseudorabies virus primary enveloped (perinuclear) C-capsid (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-17976: 
Pseudorabies nuclear C-capsids (US3 KO) vertices determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18473: 
Subtomogram average of pseudorabies virus nuclear egress complex helical form (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18474: 
Subtomogram average of pseudorabies virus nuclear egress complex (UL31/34) determined in situ
Method: subtomogram averaging / : Prazak V, Grange M, Vasishtan D

EMDB-18479: 
Pseudorabies virus cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18480: 
Pseudorabies virus nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18481: 
Herpes simplex virus 1 cytosolic C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-18483: 
Herpes simplex virus 1 nuclear C-capsid (WT) vertices determined in situ
Method: subtomogram averaging / : Mironova Y, Prazak V, Vasishtan D

EMDB-38232: 
The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8xbx: 
The cryo-EM structure of the RAD51 L2 loop bound to the linker DNA with the blunt end of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-36442: 
The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-36443: 
The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-36444: 
The cryo-EM structure of the RAD51 filament bound to the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-38228: 
The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-38229: 
The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-38230: 
The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-38231: 
The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-38233: 
The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8jnd: 
The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8jne: 
The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8jnf: 
The cryo-EM structure of the RAD51 filament bound to the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8xbt: 
The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8xbu: 
The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8xbv: 
The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8xbw: 
The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

PDB-8xby: 
The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome
Method: single particle / : Shioi T, Hatazawa S, Ogasawara M, Takizawa Y, Kurumizaka H

EMDB-19440: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP

PDB-8rqf: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP
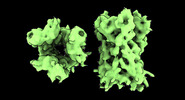
EMDB-17704: 
Subtomogram average of Vaccinia A10 trimer with open center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J
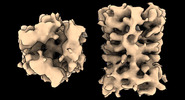
EMDB-17708: 
Subtomogram average of Vaccinia A10 trimer with tight center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-17753: 
Subtomogram average of Vaccinia A10 trimer from in situ cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-16440: 
C1 reconstruction of Tanay virus particle
Method: single particle / : Okamoto K, Song C, Miyazaki N, Murata K

EMDB-35448: 
Overlapping tri-nucleosome
Method: single particle / : Nishimura M, Fujii T, Tanaka H, Maehara K, Nozawa K, Takizawa Y, Ohkawa Y, Kurumizaka H

PDB-8ihl: 
Overlapping tri-nucleosome
Method: single particle / : Nishimura M, Fujii T, Tanaka H, Maehara K, Nozawa K, Takizawa Y, Ohkawa Y, Kurumizaka H

EMDB-41138: 
CryoEM structure of MFRV-VILP bound to IGF1Rzip
Method: single particle / : Kirk NS

EMDB-42940: 
Dimer of Hendra virus prefusion F trimers
Method: single particle / : Byrne PO, Blade EG, McLellan JS

EMDB-19035: 
Composite map of the Emiliania huxleyi virus 201 (EhV-201) symmetry expanded from cryo-EM structure of virion vertex 120 nm in diameter.
Method: subtomogram averaging / : Homola M, Buttner CR, Fuzik T, Novacek J, Chaillet M, Forster F, Plevka P

EMDB-19036: 
Cryo-EM structure of the Emiliania huxleyi virus 201 (EhV-201) virion vertex with a diameter of 50 nm and a mask applied on the capsid layer.
Method: subtomogram averaging / : Homola M, Buttner CR, Fuzik T, Novacek J, Chaillet M, Forster F, Plevka P

EMDB-32974: 
Capsid structure of Staphylococcus jumbo bacteriophage S6
Method: single particle / : Koibuchi W, Uchiyama J, Matsuzaki S, Murata K, Iwasaki K, Miyazaki N

PDB-7x30: 
Capsid structure of Staphylococcus jumbo bacteriophage S6
Method: single particle / : Koibuchi W, Uchiyama J, Matsuzaki S, Murata K, Iwasaki K, Miyazaki N

EMDB-36251: 
RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H

EMDB-36252: 
RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H

EMDB-36253: 
RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H

EMDB-37848: 
RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(0) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H

PDB-8jh2: 
RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H

PDB-8jh3: 
RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H

PDB-8jh4: 
RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome
Method: single particle / : Akatsu M, Fujita R, Ogasawara M, Ehara H, Kujirai T, Takizawa Y, Sekine S, Kurumizaka H
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
