-Search query
-Search result
Showing all 32 items for (author: yeung & ho)

EMDB-33887: 
Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592)
Method: single particle / : Oh ZG, Ang WSL, Bhushan S, Mueller-Cajar O

EMDB-35158: 
Rubisco from Phaeodactylum tricornutum
Method: single particle / : Oh ZG, Ang WSL, Bhushan S, Mueller-Cajar O

EMDB-35159: 
PYCO1(452-592) from Phaeodactylum tricornutum
Method: single particle / : Oh ZG, Ang WSL, Bhushan S, Mueller-Cajar O

EMDB-35166: 
Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592)
Method: single particle / : Oh ZG, Ang WSL, Bhushan S, Mueller-Cajar O

PDB-7yk5: 
Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592)
Method: single particle / : Oh ZG, Ang WSL, Bhushan S, Mueller-Cajar O
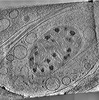
EMDB-29207: 
CryoET tomogram of mitochondria in BACHD mouse model neuron
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W
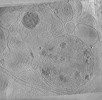
EMDB-29208: 
CryoET tomogram of BACHD mouse model neuron showing sheet aggregates
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W
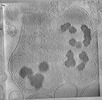
EMDB-29210: 
CryoET tomogram of purified mitochondria from HD patient iPSC-derived neuron (Q109)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
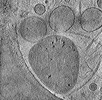
EMDB-29211: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with PIAS1 hetKO treatment
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-28668: 
CryoET tomogram of iPSC-derived control non-HD neuron (Q18)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-28944: 
CryoET tomogram of iPSC-derived control non-HD neuron (Q20)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-28946: 
CryoET tomogram of HD patient iPSC-derived neuron (Q53)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-29074: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29075: 
CryoET tomogram of HD patient iPSC-derived neuron (Q77)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29076: 
CryoET tomogram of HD patient iPSC-derived neuron (Q109)
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29079: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) showing sheet aggregate
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29080: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with PIAS1 hetKO treatment
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W
EMDB-29081: 
CryoET tomogram of HD patient iPSC-derived neuron (Q66) with GRSF1 KD treatment
Method: electron tomography / : Wu GH, Smith-Geater C, Galaz-Montoya JG, Mitchell PG, Thompson LM, Chiu W

EMDB-29083: 
CryoET tomogram of mitochondria in WT mouse neuron
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W

EMDB-29084: 
CryoET tomogram of mitochondria in BACHD-dN17 mouse model neuron
Method: electron tomography / : Wu GH, Galaz-Montoya JG, Gu Y, Mitchell PG, Wu C, Chiu W

EMDB-32497: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (focused refinement on Fab-RBD)
Method: single particle / : Zeng JW

EMDB-32498: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U)
Method: single particle / : Zeng JW, Ge JW

EMDB-32499: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d)
Method: single particle / : Zeng JW, Wang XW

PDB-7wh8: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (focused refinement on Fab-RBD)
Method: single particle / : Zeng JW

PDB-7whb: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (3U)
Method: single particle / : Zeng JW, Ge JW, Wang XQ

PDB-7whd: 
SARS-CoV-2 spike in complex with the ZB8 neutralizing antibody Fab (2u1d)
Method: single particle / : Zeng JW, Wang XW, Ge JW, Wang ZY
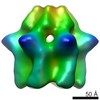
EMDB-2589: 
Inter-ring rotations of AAA ATPase p97
Method: single particle / : Yeung HO, Forster A, Bebeacua C, Niwa H, Ewens C, McKeown C, Zhang X, Freemont PS

EMDB-2590: 
Inter-ring rotations of AAA ATPase p97
Method: single particle / : Yeung HO, Forster A, Bebeacua C, Niwa H, Ewens C, McKeown C, Zhang X, Freemont PS

EMDB-2591: 
Inter-ring rotations of AAA ATPase p97
Method: single particle / : Yeung HO, Forster A, Bebeacua C, Niwa H, Ewens C, McKeown C, Zhang X, Freemont PS
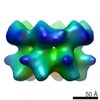
EMDB-2592: 
Inter-ring rotations of AAA ATPase p97
Method: single particle / : Yeung HO, Forster A, Bebeacua C, Niwa H, Ewens C, McKeown C, Zhang X, Freemont PS
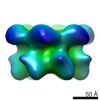
EMDB-2593: 
Inter-ring rotations of AAA ATPase p97
Method: single particle / : Yeung HO, Forster A, Bebeacua C, Niwa H, Ewens C, McKeown C, Zhang X, Freemont PS
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
