-Search query
-Search result
Showing 1 - 50 of 461 items for (author: ye & xiang)

EMDB-36651: 
hOCT1 in complex with metformin in outward open conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36652: 
hOCT1 in complex with metformin in outward occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36653: 
hOCT1 in complex with metformin in inward occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36654: 
hOCT1 in complex with nb5660 in inward facing partially open 1 conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36655: 
hOCT1 in complex with nb5660 in inward facing fully open conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36656: 
hOCT1 in complex with nb5660 in inward facing partially open 2 conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36657: 
hOCT1 in complex with spironolactone in outward facing partially occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

EMDB-36658: 
hOCT1 in complex with spironolactone in inward facing occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jts: 
hOCT1 in complex with metformin in outward open conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jtt: 
hOCT1 in complex with metformin in outward occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jtv: 
hOCT1 in complex with metformin in inward occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jtw: 
hOCT1 in complex with nb5660 in inward facing partially open 1 conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jtx: 
hOCT1 in complex with nb5660 in inward facing fully open conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jty: 
hOCT1 in complex with nb5660 in inward facing partially open 2 conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8jtz: 
hOCT1 in complex with spironolactone in outward facing partially occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X

PDB-8ju0: 
hOCT1 in complex with spironolactone in inward facing occluded conformation
Method: single particle / : Zhang S, Zhu A, Kong F, Chen J, Lan B, He G, Gao K, Cheng L, Yan C, Chen L, Liu X
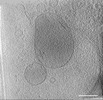
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT
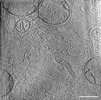
EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-38313: 
Structure of yeast replisome associated with FACT and histone hexamer, the region of FACT-Histones optimized local map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38314: 
Structure of yeast replisome associated with FACT and histone hexamer,Conformation-2
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38315: 
Structure of yeast replisome associated with FACT and histone hexamer, the region of polymerase epsilon optimized local map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38316: 
Structure of yeast replisome associated with FACT and histone hexamer, Conformation-1
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-38317: 
Structure of yeast replisome associated with FACT and histone hexamer, Composite map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

PDB-8xgc: 
Structure of yeast replisome associated with FACT and histone hexamer, Composite map
Method: single particle / : Li N, Gao Y, Yu D, Gao N, Zhai Y

EMDB-36384: 
Cryo-EM structure of the prokaryotic SPARSA system complex
Method: single particle / : Xu X, Zhen X, Long F

EMDB-36385: 
Cryo-EM structure of the prokaryotic SPARSA system complex
Method: single particle / : Xu X, Zhen X, Long F

PDB-8jkz: 
Cryo-EM structure of the prokaryotic SPARSA system complex
Method: single particle / : Xu X, Zhen X, Long F

PDB-8jl0: 
Cryo-EM structure of the prokaryotic SPARSA system complex
Method: single particle / : Xu X, Zhen X, Long F
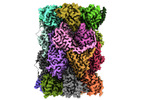
EMDB-29764: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304
Method: single particle / : Hsu HC, Li H
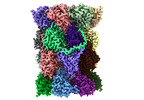
EMDB-29765: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs
Method: single particle / : Hsu HC, Li H

EMDB-42148: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304
Method: single particle / : Hsu HC, Li H

PDB-8g6e: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304
Method: single particle / : Hsu HC, Li H

PDB-8g6f: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs
Method: single particle / : Hsu HC, Li H

PDB-8ud9: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304
Method: single particle / : Hsu HC, Li H

EMDB-35932: 
Local refined cryo-EM reconstruction of Omicron BA.5 RBD in complex with 8-9D Fab
Method: single particle / : Xiang Y, Li R

EMDB-35934: 
Cryo-EM reconstruction of SARS-CoV2 Omicron BA.5 spike in complex with 8-9D Fabs
Method: single particle / : Xiang Y, Li R

PDB-8j1t: 
Local refined cryo-EM structure of Omicron BA.5 RBD in complex with 8-9D Fab
Method: single particle / : Xiang Y, Li R

PDB-8j1v: 
Cryo-EM structure of SARS-CoV2 Omicron BA.5 spike in complex with 8-9D Fabs
Method: single particle / : Xiang Y, Li R

EMDB-34449: 
Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 tetrameric complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34450: 
Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 hexameric complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34451: 
Cryo-EM Structure of the KBTBD2-Cul3-Rbx1 octameric complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34452: 
Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a dimeric complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34453: 
Cryo-EM Structure of the KBTBD2-CUL3-Rbx1-p85a tetrameric complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34454: 
local refinement of KBTBD2-p85a: KBTBD2-CRL3-p85a complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34455: 
Cryo-EM Structure of the KBTBD2-CRL3~N8-CSN(mutate) complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L

EMDB-34456: 
local refinement of CSN1/2/4/5/6-CRL3/CTD-N8: KBTBD2-CRL3-CSN(mutate) complex
Method: single particle / : Hu Y, Mao Q, Chen Z, Sun L
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
