-Search query
-Search result
Showing 1 - 50 of 1,532 items for (author: yang & gh)

EMDB-43705: 
HIV-1 wild-type intasome core
Method: single particle / : Li M, Craigie R

EMDB-43756: 
HIV-1 P5-IN intasome core
Method: single particle / : Li M, Craigie R

EMDB-43761: 
HIV-1 intasome core assembled with wild-type integrase, 1F
Method: single particle / : Li M, Craigie R

PDB-8w34: 
HIV-1 intasome core assembled with wild-type integrase, 1F
Method: single particle / : Li M, Craigie R

EMDB-37847: 
potassium outward rectifier channel SKOR
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

EMDB-37855: 
SKOR D312N L271P double mutation
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

PDB-8wtz: 
potassium outward rectifier channel SKOR
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

PDB-8wui: 
SKOR D312N L271P double mutation
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

EMDB-41907: 
Computationally Designed, Expandable O4 Octahedral Handshake Nanocage
Method: single particle / : Weidle C, Borst A

EMDB-42031: 
Computational Designed Nanocage O43_129_+8
Method: single particle / : Weidle C, Kibler RD

EMDB-43318: 
Twistless helix 12 repeat ring design R12B
Method: single particle / : Calise SJ, Kollman JM

EMDB-29974: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

EMDB-41364: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

EMDB-42906: 
Computational Designed Nanocage O43_129
Method: single particle / : Weidle C, Kibler RD

EMDB-42944: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

PDB-8gel: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

PDB-8tl7: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

PDB-8v3b: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

EMDB-41816: 
Cryo-EM structure of the RAF1-HSP90-CDC37 complex in the closed state
Method: single particle / : Finci LI, Simanshu DK

EMDB-41817: 
Cryo-EM structure of the HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

EMDB-41818: 
Cryo-EM structure of the cross-linked HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

PDB-8u1l: 
Cryo-EM structure of the RAF1-HSP90-CDC37 complex in the closed state
Method: single particle / : Finci LI, Simanshu DK

PDB-8u1m: 
Cryo-EM structure of the HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

PDB-8u1n: 
Cryo-EM structure of the cross-linked HSP90 dimer (NTD-MD) in the semi-open state
Method: single particle / : Finci LI, Simanshu DK

EMDB-41730: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Ribavirin
Method: single particle / : Wright NJ, Lee SY
EMDB-41731: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in MSP2N2 nanodiscs, apo state
Method: single particle / : Wright NJ, Lee SY
EMDB-41732: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, consensus reconstruction
Method: single particle / : Wright NJ, Lee SY
EMDB-41733: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with GS-441524, subset reconstruction
Method: single particle / : Wright NJ, Lee SY
EMDB-41734: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with N-hydroxycytidine
Method: single particle / : Wright NJ, Lee SY
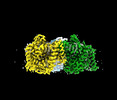
EMDB-41735: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with PSI-6206
Method: single particle / : Wright NJ, Lee SY

EMDB-41736: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-INT1 conformation
Method: single particle / : Wright NJ, Lee SY
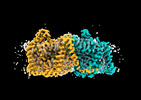
EMDB-41737: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-INT3 conformation
Method: single particle / : Wright NJ, Lee SY
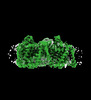
EMDB-41738: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT2-INT2 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41739: 
Cryo-EM structure of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT1-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41740: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, consensus reconstruction
Method: single particle / : Wright NJ, Lee SY

EMDB-41741: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-OFS conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41742: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41743: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-OFS-INT3 conformation
Method: single particle / : Wright NJ, Lee SY
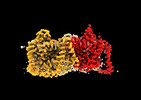
EMDB-41744: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT1-INT1-OFS conformation (from ensemble analysis)
Method: single particle / : Wright NJ, Lee SY
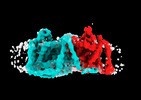
EMDB-41745: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-OFS conformation
Method: single particle / : Wright NJ, Lee SY
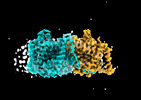
EMDB-41746: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-INT1 conformation
Method: single particle / : Wright NJ, Lee SY
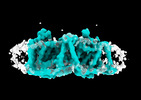
EMDB-41747: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, INT3-INT3-INT3 conformation
Method: single particle / : Wright NJ, Lee SY
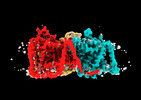
EMDB-41748: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-INT1-INT3 (clockwise) conformation
Method: single particle / : Wright NJ, Lee SY
EMDB-41749: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 1, OFS-INT1-INT3 (counterclockwise) conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41750: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT2-INT2-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41751: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, INT1-INT1-INT1 conformation
Method: single particle / : Wright NJ, Lee SY

EMDB-41752: 
Cryo-EM reconstruction of bovine concentrative nucleoside transporter 3 in complex with Molnupiravir, condition 2, consensus reconstruction
Method: single particle / : Wright NJ, Lee SY
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
