-Search query
-Search result
Showing 1 - 50 of 714 items for (author: wong & w)

EMDB-29907: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v); consensus map with only Fab 1G01 resolved
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-29908: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), locally refined map
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-29909: 
Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gat: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v), based on consensus cryo-EM map with only Fab 1G01 resolved
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gau: 
Structure of human NDS.1 Fab and 1G01 Fab in complex with influenza virus neuraminidase from A/Indiana/10/2011 (H3N2v)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

PDB-8gav: 
Structure of human NDS.3 Fab in complex with influenza virus neuraminidase from A/Darwin/09/2021 (H3N2)
Method: single particle / : Tsybovsky Y, Lederhofer J, Kwong PD, Kanekiyo M

EMDB-37631: 
Hepatitis B virus capsid (HBV core protein)
Method: single particle / : Yip RPH, Lai LTF, Lau WCY, Ngo JCK, Kwok DCY

EMDB-37634: 
SR protein kinase 2 bound at 2-fold vertex of Hepatitis B virus capsid
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK

EMDB-38062: 
Hepatitis B virus capsid in complex with SR protein kinase 2
Method: single particle / : Yip RPH, Lai LTF, Kwok DCY, Lau WCY, Ngo JCK
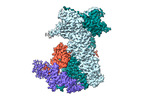
EMDB-18498: 
Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex
Method: single particle / : Kwong HS, Grandvuillemin L, Sirounian S, Ancelin A, Lai-Kee-Him J, Carivenc C, Lancey C, Ragan TJ, Hesketh EL, Bourguet W, Gruszczyk J

PDB-8qmo: 
Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex
Method: single particle / : Kwong HS, Grandvuillemin L, Sirounian S, Ancelin A, Lai-Kee-Him J, Carivenc C, Lancey C, Ragan TJ, Hesketh EL, Bourguet W, Gruszczyk J

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

EMDB-36887: 
Structure of GPR34-Gi complex
Method: single particle / : Wong TS, Zeng ZC, Xiong TT, Gan SY, He GD, Du Y

PDB-8k4n: 
Structure of GPR34-Gi complex
Method: single particle / : Wong TS, Zeng ZC, Xiong TT, Gan SY, He GD, Du Y
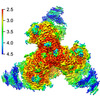
EMDB-28617: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB
Method: single particle / : Pletnev S, Kwong P

EMDB-28618: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB
Method: single particle / : Pletnev S, Kwong P

EMDB-28619: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB
Method: single particle / : Pletnev S, Kwong P

PDB-8euu: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB
Method: single particle / : Pletnev S, Kwong P

PDB-8euv: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB
Method: single particle / : Pletnev S, Kwong P

PDB-8euw: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB
Method: single particle / : Pletnev S, Kwong P

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD

EMDB-28115: 
Western Equine Encephalitis Virus-Like Particle in Complex with SKW19 Fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong P

EMDB-28116: 
Western Equine Encephalitis Virus-Like Particle in Complex with SKW24 Fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong PD

EMDB-28117: 
Eastern Equine Encephalitis Virus-Like Particle in Complex with SKE26 Fab
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

EMDB-37096: 
Rpd3S histone deacetylase complex
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, Jun H

EMDB-37122: 
Rpd3S in complex with 187bp nucleosome
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37123: 
Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37124: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37125: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2
Method: single particle / : Dong S, Li H, Meilin W, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37126: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

EMDB-37127: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Zhang Y, Wong KH, Chang X, Chao WCH, He J

PDB-8kc7: 
Rpd3S histone deacetylase complex
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

PDB-8kd2: 
Rpd3S in complex with 187bp nucleosome
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

PDB-8kd3: 
Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

PDB-8kd4: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

PDB-8kd5: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Chang X, Chao WCH, He J

PDB-8kd6: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Zhang X, Chao WCH, He J

PDB-8kd7: 
Rpd3S in complex with nucleosome with H3K36MLA modification and 167bp DNA
Method: single particle / : Dong S, Li H, Wang M, Rasheed N, Zou B, Gao X, Guan J, Li W, Zhang J, Wang C, Zhou N, Shi X, Li M, Zhou M, Huang J, Li H, Zhang Y, Wong KH, Chang X, Chao WCH, He J

EMDB-41302: 
Lassa GPC trimer in complex with Fab GP23
Method: single particle / : Gorman J, Kwong PD

EMDB-28891: 
cryo-EM structure of a structurally designed Human metapneumovirus F protein in complex with antibody MPE8
Method: single particle / : Zhou T, Kwong PD, Morano NC, Ou L

PDB-8f6x: 
cryo-EM structure of a structurally designed Human metapneumovirus F protein in complex with antibody MPE8
Method: single particle / : Zhou T, Kwong PD, Morano NC, Ou L
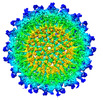
EMDB-28118: 
Cryo-EM structure of Antibody SKW11 in complex with Western Equine Encephalitis Virus-Like Particle
Method: single particle / : Cerutti G, Verardi R, Roederer M, Shapiro L
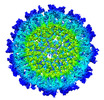
EMDB-28119: 
Cryo-EM structure of Antibody SKT05 in complex with Western Equine Encephalitis Virus-like Particle
Method: single particle / : Cerutti G, Verardi R, Roederer M, Shapiro L

EMDB-28187: 
Cryo-EM I1 reconstruction of antibody SKV09 in complex with VEEV alphavirus VLP
Method: single particle / : Casner RG, Verardi R, Roederer M, Shapiro L

EMDB-28188: 
Cryo-EM I1 reconstruction of antibody SKV16 in complex with VEEV alphavirus VLP
Method: single particle / : Casner RG, Verardi R, Roederer M, Shapiro L

EMDB-27757: 
Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab
Method: single particle / : Pletnev S, Verardi R, Roedeger M, Kwong P

EMDB-28056: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD

EMDB-28058: 
Venezuelan equine encephalitis virus-like particle in complex with Fab SKT05, local refinement
Method: single particle / : Tsybovsky Y, Pletnev S, Verardi R, Roederer M, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
