-Search query
-Search result
Showing all 18 items for (author: witte & g)

EMDB-13108: 
High resolution structure of cytochrome bd-II oxidase from E. coli
Method: single particle / : Grund TN, Wu D, Bald D, Michel H, Safarian S

EMDB-13117: 
High resolution structure of cytochrome bd-II oxidase from E. coli at 2.46 A resolution
Method: single particle / : Grund TN, Wu D, Bald D, Michel H, Safarian S

EMDB-13121: 
High resolution structure of cytochrome bd-II oxidase from E. coli at 2.65 A resolution
Method: single particle / : Grund TN, Wu D, Bald D, Michel H, Safarian S

EMDB-13122: 
High resolution structure of cytochrome bd-II oxidase from E. coli at 2.55 A resolution
Method: single particle / : Grund TN, Wu D, Bald D, Michel H, Safarian S

PDB-7oy2: 
High resolution structure of cytochrome bd-II oxidase from E. coli
Method: single particle / : Grund TN, Wu D, Bald D, Michel H, Safarian S

EMDB-12051: 
S. agalactiae BusR in complex with its busAB-promotor DNA
Method: single particle / : Bandera AM, Witte G

EMDB-13119: 
S. agalactiae BusR in complex with its busA-promotor DNA
Method: single particle / : Bandera AM, Witte G

PDB-7b5y: 
S. agalactiae BusR in complex with its busAB-promotor DNA
Method: single particle / : Bandera AM, Witte G

PDB-7oz3: 
S. agalactiae BusR in complex with its busA-promotor DNA
Method: single particle / : Bandera AM, Witte G

EMDB-11601: 
CryoEM Structure of cGAS Nucleosome complex
Method: single particle / : Michalski S, de Oliveira Mann CC, Witte G, Bartho J, Lammens K, Hopfner KP

PDB-7a08: 
CryoEM Structure of cGAS Nucleosome complex
Method: single particle / : Michalski S, de Oliveira Mann CC, Witte G, Bartho J, Lammens K, Hopfner KP

EMDB-21005: 
Negative stain EM map of CODH/ACS in the closed conformation
Method: single particle / : Cohen SE, Brignole EJ, Drennan CL
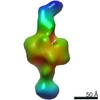
EMDB-21006: 
Negative stain EM map of CODH/ACS in the hyperextended conformation
Method: single particle / : Cohen SE, Brignole EJ, Drennan CL

EMDB-21007: 
Negative stain EM map of CODH/ACS in the half-extended conformation
Method: single particle / : Cohen SE, Brignole EJ, Drennan CL
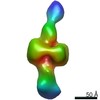
EMDB-21008: 
Negative stain EM map of CODH/ACS in the extended conformation
Method: single particle / : Cohen SE, Brignole EJ, Drennan CL

PDB-4v6t: 
Structure of the bacterial ribosome complexed by tmRNA-SmpB and EF-G during translocation and MLD-loading
Method: single particle / : Ramrath DJF, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CMT

EMDB-2179: 
Electron cryo-microscopy of C. thermophilum RAC bound to the 80S ribosome
Method: single particle / : Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I

EMDB-5386: 
tmRNA translocation and MLD-loading on the ribosome: a 70S-tmRNA-EF-G complex
Method: single particle / : Ramrath DJF, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CMT
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
