-Search query
-Search result
Showing 1 - 50 of 185 items for (author: william & r & schief)
EMDB-40242: 
BG505 MD39 SOSIP in complex with Rh.NJ85 wk12 gp120GH, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40243: 
BG505 MD39 SOSIP in complex with Rh.NJ86 wk12 V1V3, C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40244: 
BG505 MD39 SOSIP in complex with Rh.NJ76 wk12 C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB
EMDB-40252: 
BG505 MD39 SOSIP in complex with Rh.NK04 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40254: 
BG505 MD39 SOSIP in complex with Rh.NJ75 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40255: 
BG505 MD39 SOSIP in complex with Rh.NJ87 wk12 C3V5 and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40256: 
BG505 MD39 SOSIP in complex with Rh.NJ84 wk12 V1V3, gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40257: 
BG505 MD39 SOSIP in complex with Rh.NJ77 wk12 V1V3, C3V5, N611/FP and base epitope polyclonal
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-25622: 
Negative Stain EM reconstruction of ApexGT3 in complex with two copies of PG9 Fab
Method: single particle / : Berndsen ZT

EMDB-27610: 
BG505 MD39 SOSIP in complex with Rh.NJ82 wk13 N611 and base epitope pAbs
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
EMDB-27611: 
BG505 MD39 SOSIP in complex with Rh.NJ95 wk13 base epitope pAb
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
EMDB-27612: 
BG505 MD39 SOSIP in complex with Rh.NK05 wk13 base epitope pAb
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB
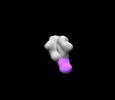
EMDB-27614: 
BG505 MD39 SOSIP in complex with Rh.NJ79 wk13 base epitope pAb
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB

EMDB-27615: 
BG505 MD39 SOSIP in complex with Rh.NJ93 wk13 base and V5/C3 epitope pAbs
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB

EMDB-25732: 
HIV-1 Envelope ApexGT2.2MUT in complex with PCT64.LMCA Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25733: 
HIV-1 Envelope ApexGT2 in complex with PCT64.35S Fab and RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25734: 
HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25735: 
HIV-1 Envelope ApexGT3 in complex with PG9.iGL Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25736: 
HIV-1 Envelope ApexGT3.N130 in complex with PG9 Fab
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t73: 
HIV-1 Envelope ApexGT2.2MUT in complex with PCT64.LMCA Fab
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t74: 
HIV-1 Envelope ApexGT2 in complex with PCT64.35S Fab and RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t75: 
HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t76: 
HIV-1 Envelope ApexGT3 in complex with PG9.iGL Fab
Method: single particle / : Berndsen ZT, Ward AB

PDB-7t77: 
HIV-1 Envelope ApexGT3.N130 in complex with PG9 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-27036: 
N332-GT2 trimer in complex with pooled polyclonal fab from BG18-exposed mice (day 14)
Method: single particle / : Jackson AM, Ward AB

EMDB-27037: 
N332-GT2 trimer in complex with pooled polyclonal fab from wild type mice (day 42)
Method: single particle / : Jackson AM, Torres JL, Ward AB

EMDB-27038: 
N332-GT2 trimer in complex with pooled polyclonal fab from BG18-exposed mice (day 42)
Method: single particle / : Jackson AM, Nogal B, Ward AB

EMDB-27039: 
SARS-CoV-2 S protein mix in complex with Novavax NHP 36057 polyclonal Fab
Method: single particle / : Jackson AM, Bangaru S, Ward AB

EMDB-27040: 
SARS-CoV-2 S protein mix in complex with Novavax NHP 36964 polyclonal Fab
Method: single particle / : Jackson AM, Bangaru S, Ward AB

EMDB-27041: 
SARS-CoV-2 S protein mix in complex with Novavax NHP 37452 polyclonal Fab
Method: single particle / : Jackson AM, Bangaru S, Ward AB

EMDB-27042: 
SARS-CoV-2 S protein mix in complex with Novavax NHP 37611 polyclonal Fab
Method: single particle / : Jackson AM, Bangaru S, Ward AB

EMDB-27197: 
SARS-CoV-2 spike protein complexed with polyclonal Fab from donor D (Donor Lotus-25)
Method: single particle / : Bangaru S, Jackson AM, Ward AB

EMDB-27198: 
SARS-CoV-2 spike protein complexed with polyclonal Fab from donor C (Donor 1992)
Method: single particle / : Bangaru S, Sewall LM, Ward AB

EMDB-27199: 
SARS-CoV-2 spike protein complexed with polyclonal Fab from donor B (Donor 1989)
Method: single particle / : Bangaru S, Sewall LM, Ward AB

EMDB-27200: 
SARS-CoV-2 spike protein complexed with polyclonal Fab from donor A (Donor 1988)
Method: single particle / : Bangaru S, Sewall LM, Ward AB

EMDB-27601: 
BG505 MD39 SOSIP in complex with Rh.K409 wk13 base, FP/gp41, V5/C3 and N611 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27602: 
BG505 MD39 SOSIP in complex with Rh.K487 wk13 base, FP/gp41, and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB
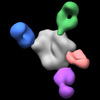
EMDB-27603: 
BG505 MD39 SOSIP in complex with Rh.DGT5 wk13 base, N335/N289, V1/V3 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB
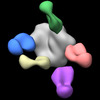
EMDB-27604: 
BG505 MD39 SOSIP in complex with Rh.K949 wk33 base, N335/N289, V1/V3, N611 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27605: 
BG505 MD39 SOSIP in complex with Rh.L282 wk33 base, N335/N289, FP/gp41 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27607: 
BG505 MD39 SOSIP in complex with Rh.DHHT wk33 base, FP/gp41 and V5/C3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27608: 
BG505 MD39 SOSIP in complex with Rh.DHID wk33 base, N355/N289 and V1/V3 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-27609: 
BG505 MD39 SOSIP in complex with Rh.NJ78 wk13 V5/C3 and base epitope pAbs
Method: single particle / : Torres JL, Lee WH, Ozorowski G, Ward AB

EMDB-27600: 
BG505 MD39 SOSIP in complex with Rh.L614 wk13 base, N335/N289, V5/C3 and N611 epitope pAbs
Method: single particle / : Sewall LM, Richey ST, Ozorowski G, Ward AB

EMDB-23778: 
Negative stain EM map of BG505 SOSIP in complex with Rh4O9.8 monoclonal antibody (as Fab fragment)
Method: single particle / : Antanasijevic A, Ward AB

EMDB-23779: 
BG505 SOSIP.v5.2 in complex with the polyclonal Fab pAbC-1 from animal Rh.4O9
Method: single particle / : Antanasijevic A, Ozorowski G, Nogal B, Ward AB

EMDB-23780: 
BG505 SOSIP MD39 in complex with the monoclonal antibodies Rh.33104 mAb.1 and RM20A3
Method: single particle / : Antanasijevic A, Ward AB

EMDB-23801: 
BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh.33172 mAb.1 and RM19R
Method: single particle / : Antanasijevic A, Ward AB

PDB-7mdt: 
BG505 SOSIP.v5.2 in complex with the monoclonal antibody Rh4O9.8 (as Fab fragment)
Method: single particle / : Antanasijevic A, Ozorowski G, Nogal B, Ward AB

PDB-7mdu: 
BG505 SOSIP MD39 in complex with the monoclonal antibodies Rh.33104 mAb.1 and RM20A3
Method: single particle / : Antanasijevic A, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
