-Search query
-Search result
Showing all 40 items for (author: william & c & olson)

EMDB-16540: 
Neurofascin isoform NF155 extracellular domain
Method: single particle / : McKie SJ, Deane JE, Butt BG

EMDB-27221: 
Cryo-EM map of human LIF signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27227: 
Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from selected full particle dataset, with better resolution in the interaction core region
Method: single particle / : Zhou Y, Franklin MC

EMDB-27228: 
Cryo-EM structure of human CNTFR alpha in complex with the Fab fragments of two antibodies
Method: single particle / : Zhou Y, Franklin MC

EMDB-27229: 
Cryo-EM map of human CNTF signaling complex with full extracellular domains: refined from a particle subset, with lower global resolution but improved LIFR D6-D8 density
Method: single particle / : Zhou Y, Franklin MC

EMDB-27230: 
Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha
Method: single particle / : Zhou Y, Franklin MC

EMDB-27231: 
Cryo-EM map of human CLCF1 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27244: 
Cryo-EM map of detergent-solubilized human IL-6 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27246: 
Cryo-EM map of human IL-27 signaling complex with full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

EMDB-27247: 
Cryo-EM map of human IL-27 signaling complex: focused refinement on the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d6a: 
Cryo-EM structure of human LIF signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d74: 
Cryo-EM structure of human CNTF signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d7e: 
Cryo-EM structure of human CNTFR alpha in complex with the Fab fragments of two antibodies
Method: single particle / : Zhou Y, Franklin MC

PDB-8d7h: 
Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha
Method: single particle / : Zhou Y, Franklin MC

PDB-8d7r: 
Cryo-EM structure of human CLCF1 signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC

PDB-8d82: 
Cryo-EM structure of human IL-6 signaling complex in detergent: model containing full extracellular domains
Method: single particle / : Zhou Y, Franklin MC

PDB-8d85: 
Cryo-EM structure of human IL-27 signaling complex: model containing the interaction core region
Method: single particle / : Zhou Y, Franklin MC
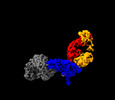
EMDB-28523: 
Structure of interleukin receptor common gamma chain (IL2Rgamma) in complex with two antibodies
Method: single particle / : Franklin MC, Romero Hernandez A

PDB-8epa: 
Structure of interleukin receptor common gamma chain (IL2Rgamma) in complex with two antibodies
Method: single particle / : Franklin MC, Romero Hernandez A

EMDB-24073: 
Complex of Bet v 1 with the Fab fragments of a three antibody cocktail
Method: single particle / : Romero-Hernandez A, Franklin MC

PDB-7mxl: 
Complex of Bet v 1 with the Fab fragments of a three antibody cocktail
Method: single particle / : Romero-Hernandez A, Franklin MC

EMDB-22137: 
Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies
Method: single particle / : Franklin MC, Saotome K, Romero Hernandez A, Zhou Y

PDB-6xdg: 
Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies
Method: single particle / : Franklin MC, Saotome K, Romero Hernandez A, Zhou Y

EMDB-7900: 
REGN3479 antibody Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Ward AB

EMDB-7901: 
REGN3470 antibody Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Ward AB

EMDB-7902: 
REGN3471 antibody Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Ward AB

EMDB-8141: 
Cryo-EM structure of the human FANCD2/FANCI complex reveals a novel Tower domain required for FANCD2 monoubiquitination- Full length FANCD2/FANCI
Method: single particle / : Liang CC, Li Z, Nicholson WV, Venien-Bryan C, Cohn MA

EMDB-8142: 
Cryo-EM structure of the human FANCD2/FANCI complex reveals a novel Tower domain required for FANCD2 monoubiquitination- FANCD2-minusTower/FANCI complex
Method: single particle / : Liang CC, Li Z, Nicholson WV, Venien-Bryan C, Cohn MA

EMDB-1930: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 JR-FL
Method: single particle / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S

EMDB-5320: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144
Method: subtomogram averaging / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S
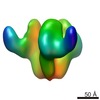
EMDB-5321: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144 with sCD4
Method: subtomogram averaging / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S

EMDB-5322: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 JR-FL with sCD4
Method: subtomogram averaging / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S

EMDB-5323: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144 with sCD4 and 17b
Method: subtomogram averaging / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S

EMDB-5324: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 JR-FL with sCD4 and 17b
Method: subtomogram averaging / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S

EMDB-5325: 
Molecular structure of soluble trimeric HIV-1 glycoprotein gp140 KNH1144 with 17b
Method: subtomogram averaging / : Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S
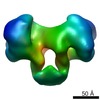
EMDB-1890: 
EcoR124 Type I DNA restriction-modification enzyme complex in closed state with bound 30bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1891: 
EcoR124 Type I DNA restriction-modification enzyme complex without DNA (open state). Low resolution 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
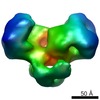
EMDB-1892: 
EcoR124 Type I DNA restriction-modification enzyme complex (in closed state) with bound DNA mimic protein Ocr from phage T7. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1893: 
EcoKI Type I DNA restriction-modification enzyme complex in closed state with bound 75bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
