-Search query
-Search result
Showing all 44 items for (author: white & jm)
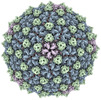
EMDB-40271: 
Mycobacterium phage Adjutor
Method: single particle / : Podgorski JM, White SJ

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
Method: single particle / : Zimmerman MI, Fremont DH
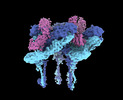
EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-28761: 
Mycobacterium phage Che8 mutant capsid (gene 110 deletion)
Method: single particle / : Podgorski JM, White SJ

EMDB-28644: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

EMDB-16268: 
E. coli BAM complex (BamABCDE) bound to darobactin B
Method: single particle / : Horne JE, Fenn KL, Radford SE, Ranson NA

EMDB-16282: 
E. coli BAM complex (BamABCDE) wild-type
Method: single particle / : Machin JM, Radford SE, Ranson NA

EMDB-40077: 
Mycobacterium phage Patience
Method: single particle / : Podgorski JM, White SJ

EMDB-27824: 
Mycobacterium phage Che8
Method: single particle / : Podgorski JM, White SJ

EMDB-27992: 
Gordonia phage Ziko
Method: single particle / : Podgorski JM, White SJ

EMDB-28012: 
Mycobacterium phage Adephagia
Method: single particle / : Podgorski JM, White SJ

EMDB-28015: 
Mycobacterium phage Bobi
Method: single particle / : Podgorski JM, White SJ

EMDB-28016: 
Arthrobacter phage Bridgette
Method: single particle / : Podgorski JM, White SJ

EMDB-28017: 
Mycobacterium phage Cain
Method: single particle / : Podgorski JM, White SJ
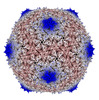
EMDB-28018: 
Gordonia phage Cozz
Method: single particle / : Podgorski JM, White SJ

EMDB-28020: 
Mycobacterium phage Ogopogo
Method: single particle / : Podgorski JM, White SJ

EMDB-28021: 
Microbacterium phage Oxtober96
Method: single particle / : Podgorski JM, White SJ

EMDB-28039: 
Mycobacteriophage Muddy capsid
Method: single particle / : Freeman KG, White SJ, Huet A, Conway JF

EMDB-24482: 
Complex III2 from Candida albicans, inhibitor free
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24483: 
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in b position
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24484: 
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in intermediate position
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24485: 
Complex III2 from Candida albicans, inhibitor free, Rieske head domain in c position
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-24486: 
Complex III2 from Candida albicans, Inz-5 bound
Method: single particle / : Di Trani JM, Rubinstein JL

EMDB-12232: 
Lateral-closed conformation of the lid-locked BAM complex (BamA E435C S665C, BamBDCE) by cryoEM
Method: single particle / : Haysom SF

EMDB-12262: 
Lateral-open conformation of the lid-locked BAM complex (BamA E435C S665C, BamBDCE) by cryoEM
Method: single particle / : Haysom SF

EMDB-12263: 
structure of a POTRA-locked BAM complex in a lateral-open conformation
Method: single particle / : Machin JM, Haysom SF

EMDB-12271: 
Lateral-open conformation of the lid-locked BAM complex (BamA E435C S665C, BamBDCE) bound by a bactericidal Fab fragment
Method: single particle / : Machin JM, Haysom SF

EMDB-12272: 
lateral-open conformation of the wild-type BAM complex (BamABCDE) bound to a bactericidal Fab fragment
Method: single particle / : Iadanza MG

EMDB-22829: 
Human Tom70 in complex with SARS CoV2 Orf9b
Method: single particle / : QCRG Structural Biology Consortium

EMDB-21122: 
Actinobacteriophage Rosebush
Method: single particle / : Podgorski JM, Calabrese J, Alexandrescu L, Jacobs-Sera D, Pope W, Hatfull G, White SJ

EMDB-21123: 
Actinobacteriophage Patience
Method: single particle / : Podgorski JM, Calabrese J, Alexandrescu L, Jacobs-Sera D, Pope W, Hatfull G, White SJ

EMDB-21124: 
Actinobacteriophage Myrna
Method: single particle / : Podgorski JM, Calabrese J, Alexandrescu L, Jacobs-Sera D, Pope W, Hatfull G, White SJ

EMDB-8225: 
Tomographic subvolume average of Ebola (EBOV-Makona) glycoprotein on the surface of virus-like particles
Method: subtomogram averaging / : Tran EEH, Subramaniam S

EMDB-8226: 
Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c13C6 antibody
Method: subtomogram averaging / : Tran EEH, Subramaniam S
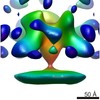
EMDB-8227: 
Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoprotein bound to c2G4 antibody
Method: subtomogram averaging / : Tran EEH, Subramaniam S

EMDB-8228: 
Tomographic subvolume average of membrane-bound Ebola (EBOV-Makona) glycoproteins bound to the chimerized human monoclonal antibody, c4G7
Method: subtomogram averaging / : Tran EEH, Subramaniam S

EMDB-6003: 
Cryo-electron tomography of full-length glycoprotein from Ebola virus-like particles
Method: subtomogram averaging / : Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S

EMDB-6004: 
Cryo-electron tomography of glycoprotein lacking the mucin-like domain from Ebola virus-like particles
Method: subtomogram averaging / : Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S
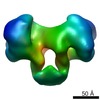
EMDB-1890: 
EcoR124 Type I DNA restriction-modification enzyme complex in closed state with bound 30bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1891: 
EcoR124 Type I DNA restriction-modification enzyme complex without DNA (open state). Low resolution 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF
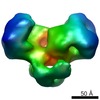
EMDB-1892: 
EcoR124 Type I DNA restriction-modification enzyme complex (in closed state) with bound DNA mimic protein Ocr from phage T7. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1893: 
EcoKI Type I DNA restriction-modification enzyme complex in closed state with bound 75bp cognate DNA fragment. 3D reconstruction by single particle analysis from negative stain EM.
Method: single particle / : Kennaway CK, Taylor JE, Song CF, Potrzebowski W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, Roberts GA, Bujnicki JM, Trinick J, Kneale GG, Dryden DTF

EMDB-1534: 
EcoKI type I RM methyltransferase with DNA mimic Ocr. Negative stain 3D.
Method: single particle / : Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DTF
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
