-Search query
-Search result
Showing 1 - 50 of 334 items for (author: white & he)

EMDB-42681: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42682: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42683: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42800: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42833: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42835: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42846: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42847: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42849: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 1 from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42856: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 2 from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42858: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

EMDB-42874: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand activated by the C1-domain of cardiac myosin binding protein C
Method: single particle / : Galkin VE, Risi CM

PDB-8uww: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uwx: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uwy: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uyd: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uz5: 
The structure of the native cardiac thin filament troponin core in Ca2+-free rotated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uz6: 
The structure of the native cardiac thin filament troponin core in Ca2+-free tilted state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uzx: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8uzy: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the upper strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v01: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 1 from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v0i: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound fully activated state 2 from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v0k: 
The structure of the native cardiac thin filament troponin core in Ca2+-bound partially activated state from the lower strand
Method: single particle / : Galkin VE, Risi CM

PDB-8v0y: 
The structure of the native cardiac thin filament troponin core in Ca2+-free state from the upper strand activated by the C1-domain of cardiac myosin binding protein C
Method: single particle / : Galkin VE, Risi CM
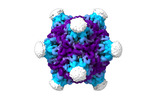
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
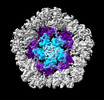
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
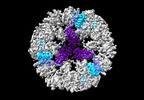
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
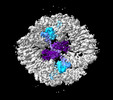
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb3: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-43013: 
Myxococcus xanthus HEnc-K417N(A) protein shell with icosahedral T=1 symmetry
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43016: 
Myxococcus xanthus HEnc-K417N(A) protein shell with tetrahedral symmetry (12 pentamers, 4 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43037: 
Myxococcus xanthus HEnc-K417N(A) protein shell with D3 symmetry (12 pentamers, 3 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43038: 
Myxococcus xanthus HEnc-K417N(A) protein shell with D6 symmetry (12 pentamers, 8 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43039: 
Myxococcus xanthus HEnc-K417N(A) protein shell with C2 symmetry (12 pentamers, 9 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43040: 
Myxococcus xanthus HEnc-K417N(A) protein shell with D3 symmetry (12 pentamers, 11 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43041: 
Myxococcus xanthus HEnc-K417N(A) protein shell with D2 symmetry (12 pentamers, 12 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43042: 
Myxococcus xanthus HEnc-K417N(A) protein shell with D2 symmetry (12 pentamers, 14 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43043: 
Myxococcus xanthus HEnc-K417N(A) protein shell with D5 symmetry (12 pentamers, 15 hexamers)
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43113: 
Myxococcus xanthus EncA WT protein shell with icosahedral symmetry T=3
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-43036: 
Myxococcus xanthus HEnc-K417N(A) protein shell with icosahedral T=3 symmetry
Method: single particle / : Hernandez C, Jenkins MC, Kopylov M

EMDB-40711: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID)

PDB-8sqn: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID)

EMDB-27272: 
CryoEM structure of Western equine encephalitis virus VLP
Method: single particle / : Zimmerman MI, Fremont DH
EMDB-27271: 
CryoEM structure of Western equine encephalitis virus VLP in complex with the avian MXRA8 receptor
Method: single particle / : Zimmerman MI, Fremont DH

EMDB-17613: 
10-mer ring of human metapneumovirus (HMPV) N-RNA
Method: single particle / : Whitehead JD, Decool H, Leyrat C, Carrique L, Fix J, Eleouet JF, Galloux M, Renner M

EMDB-17614: 
11-mer ring of human metapneumovirus (HMPV) N-RNA
Method: single particle / : Whitehead JD, Decool H, Leyrat C, Carrique L, Fix J, Eleouet JF, Galloux M, Renner M

EMDB-17615: 
Spiral of assembled human metapneumovirus (HMPV) N-RNA
Method: single particle / : Whitehead JD, Decool H, Leyrat C, Carrique L, Fix J, Eleouet JF, Galloux M, Renner M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
