-Search query
-Search result
Showing 1 - 50 of 105 items for (author: weiss & gl)

EMDB-18953: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18954: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M
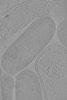
EMDB-18955: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18957: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18958: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18960: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18961: 
Cryo-electron tomogram of mechanically cryo-milled Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18962: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M
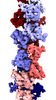
EMDB-18970: 
Subtomogram average of M66 filaments in Yersinia entomophaga cells
Method: subtomogram averaging / : Feldmueller M, Afanasyev P, Pilhofer M
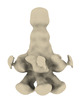
EMDB-18971: 
Subtomogram average of YenTc-Chi2-sfGFP from Yersinia entomophaga chi2-sfGFP
Method: subtomogram averaging / : Feldmueller M, Pilhofer M
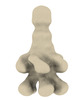
EMDB-18972: 
Subtomogram average of YenTc from Yersinia entomophaga MH96
Method: subtomogram averaging / : Feldmueller M, Pilhofer M

EMDB-19370: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19371: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19372: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19373: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19374: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19375: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19376: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19377: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19378: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19379: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19380: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19381: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-16012: 
Subtomogram average of septal junctions from Nostoc PCC 7120 in the open state
Method: subtomogram averaging / : Kieninger A, Tokarz P, Janovic A, Pilhofer M, Weiss G, Maldener I

EMDB-16033: 
Subtomogram average of septal junctions from Nostoc PCC 7120 in the closed state
Method: subtomogram averaging / : Kieninger A, Tokarz P, Janovic A, Pilhofer M, Weiss G, Maldener I

EMDB-16034: 
Subtomogram average of septal junctions from sepN mutant
Method: subtomogram averaging / : Kieninger A, Tokarz P, Janovic A, Pilhofer M, Weiss G, Maldener I

EMDB-16053: 
Subtomogram average of septal junctions from sepN-sfgfp mutant
Method: subtomogram averaging / : Kieninger A, Tokarz P, Janovic A, Pilhofer M, Weiss G, Maldener I

EMDB-16054: 
Subtomogram average of septal junctions from sepN-mbp-sfgfp mutant
Method: subtomogram averaging / : Kieninger A, Tokarz P, Janovic A, Pilhofer M, Weiss G, Maldener I

EMDB-16061: 
Electron cryotomogram of septal region from FIB milled filament of Nostoc sp. PCC 7120 sepN-mbp-sfgfp mutant
Method: electron tomography / : Kieninger A, Tokarz P, Janovic A, Pilhofer M, Weiss G, Maldener I

EMDB-11953: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-11954: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14810: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14811: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-23480: 
Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-7lpn: 
Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-12029: 
Cryo-EM structure of the contractile injection system base plate from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12030: 
Cryo-EM structure of the contractile injection system base plate from Anabaena PCC7120 (C6)
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12031: 
Cryo-EM structure of the contractile injection system base plate connector from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12032: 
Cryo-EM structure of the contractile injection system crown trimer from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12033: 
Cryo-EM structure of the contractile injection system base plate in contracted form from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-12034: 
Cryo-EM structure of the contractile injection system cap complex from Anabaena PCC7120
Method: single particle / : Eisenstein F, Weiss GL, Pilhofer M

EMDB-13770: 
Cryo-electron tomogram of cryo-FIB milled Anabaena PCC 7120
Method: electron tomography / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13771: 
Cryo-electron tomogram of cryo-FIB milled Anabaena PCC 7120, cell periphery
Method: electron tomography / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13772: 
Cryo-electron tomogram of cryo-FIB milled Anabaena PCC 7120 ghost cell
Method: electron tomography / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13773: 
Subtomogram average of baseplate region of contractile injection systems in Anabaena PCC 7120 in extended state
Method: subtomogram averaging / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13774: 
Subtomogram average of baseplate region of contractile injection systems in Anabaena PCC 7120 ghost cells in extended state
Method: subtomogram averaging / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-13775: 
Subtomogram average of baseplate region of contractile injection systems in Anabaena PCC 7120 ghost cells in contracted state
Method: subtomogram averaging / : Weiss GL, Eisenstein F, Pilhofer M

EMDB-21705: 
PL-bound rat TRPV2 in nanodiscs
Method: single particle / : Pumroy RP, Moiseenkova-Bell VY

PDB-6wkn: 
PL-bound rat TRPV2 in nanodiscs
Method: single particle / : Pumroy RP, Moiseenkova-Bell VY

EMDB-10694: 
Structure of human cGAS (K394E) bound to the nucleosome
Method: single particle / : Pathare GR, Cavadini S, Kempf G, Thoma NH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
