-Search query
-Search result
Showing 1 - 50 of 640 items for (author: way & m)

EMDB-41672: 
ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-41673: 
ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8twv: 
ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8twz: 
ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL
EMDB-42286: 
T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
EMDB-42355: 
T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
EMDB-42382: 
T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
EMDB-42390: 
T33-ml23 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

PDB-8ui2: 
T33-ml28 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

PDB-8ukm: 
T33-ml30 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

PDB-8umr: 
T33-ml35 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

PDB-8un1: 
T33-ml23 Assembly Intermediate - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
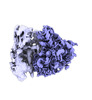
EMDB-40665: 
Puromycin reveals a distinct conformation of neuronal ribosomes.
Method: single particle / : Anadolu M, Sun J, Tian-Yi-Li J, Graber T, Ortega J, Sossin WS
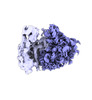
EMDB-40666: 
Class2 80S with PP tRNA
Method: single particle / : Anadolu M, Sun J, Tian-Yi-Li J, Graber T, Ortega J, Sossin WS

EMDB-40667: 
Class3 80S with broken density in P site
Method: single particle / : Anadolu M, Sun J, Tian-Yi-Li J, Graber T, Ortega J, Sossin WS
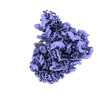
EMDB-40668: 
Class4 80S with only 60S
Method: single particle / : Ortega J

EMDB-43542: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation
Method: single particle / : Petroff II JT, Deng Z, Rau MJ, Fitzpatrick JAJ, Yuan P, Cheng WWL

PDB-8vuw: 
ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation
Method: single particle / : Petroff II JT, Deng Z, Rau MJ, Fitzpatrick JAJ, Yuan P, Cheng WWL

PDB-8t1n: 
Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold
Method: electron crystallography / : Miller JE, Cascio D, Sawaya MR, Cannon KA, Rodriguez JA, Yeates TO

EMDB-17557: 
Cryo-EM structure of cortactin-stabilized Arp2/3-complex nucleated actin branches-Local refined map on mother filament
Method: single particle / : Liu T, Moores CA
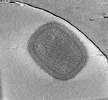
EMDB-18916: 
Cryotomogram of mature Vaccinia virus (WR) virion
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18917: 
Subtomogram average of the Vaccinia virus (WR) portal complex in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18918: 
Subtomogram average of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

PDB-8r5i: 
In situ structure of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions by flexible fitting into a cryoET map
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-17553: 
Cryo-EM structure of cortactin-stabilized Arp2/3 complex nucleated actin branches-Daughter filament consensus map
Method: single particle / : Liu T, Moores CA

EMDB-17554: 
Cryo-EM structure of cortactin-stabilized Arp2/3 nucleated actin branches-Local refined map on Arp2/3 complex
Method: single particle / : Liu T, Moores CA

EMDB-17555: 
Cryo-EM structure of cortactin-stabilized Arp2/3-complex nucleated actin branches-Local refined map on the daughter filament and cortactin density
Method: single particle / : Liu T, Moores CA

EMDB-17556: 
Cryo-EM structure of cortactin-stabilized Arp2/3-complex nucleated actin branches-Local refined map on capping protein
Method: single particle / : Liu T, Moores CA

EMDB-17558: 
Cryo-EM structure of cortactin stabilized Arp2/3-complex nucleated actin branches
Method: single particle / : Liu T, Moores CA

PDB-8p94: 
Cryo-EM structure of cortactin stabilized Arp2/3-complex nucleated actin branches
Method: single particle / : Liu T, Moores CA

EMDB-41195: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) G269V mutant, amyloid fiber
Method: helical / : Rosenberg GM, Sawaya MR, Boyer DR, Ge P, Abskharon R, Eisenberg DS
EMDB-41198: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber
Method: helical / : Rosenberg GM, Sawaya MR, Boyer DR, Ge P, Abskharon R, Eisenberg DS

PDB-8teq: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) G269V mutant, amyloid fiber
Method: helical / : Rosenberg GM, Sawaya MR, Boyer DR, Ge P, Abskharon R, Eisenberg DS

PDB-8ter: 
Tropomyosin-receptor kinase fused gene protein (TRK-fused gene protein; TFG) Low Complexity Domain (residues 237-327) P285L mutant, amyloid fiber
Method: helical / : Rosenberg GM, Sawaya MR, Boyer DR, Ge P, Abskharon R, Eisenberg DS

EMDB-41171: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, wild-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L

EMDB-41172: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, variant-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L
PDB-8tdn: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, wild-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L
PDB-8tdo: 
Cryo-EM structure of cardiac amyloid fibril from a variant ATTR I84S amyloidosis patient-3, variant-type morphology
Method: helical / : Nguyen BA, Singh V, Saelices L

EMDB-42181: 
T33-ml23 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO
EMDB-42381: 
T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

PDB-8uf0: 
T33-ml23 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

PDB-8ump: 
T33-ml35 - Designed Tetrahedral Protein Cage Using Machine Learning Algorithms
Method: single particle / : Castells-Graells R, Meador K, Sawaya MR, Yeates TO

EMDB-28829: 
ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-28830: 
ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-28831: 
ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

EMDB-28832: 
Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f32: 
ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f33: 
ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f34: 
ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL

PDB-8f35: 
Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG
Method: single particle / : Dalal V, Arcario MJ, Petroff II JT, Deitzen NM, Tan BK, Brannigan G, Cheng WWL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
