-Search query
-Search result
Showing all 46 items for (author: walker & ml)

EMDB-27232: 
Subtomogram of Fluad vaccine hemagglutinin spiked nanodisc
Method: electron tomography / : Gallagher JR, Audray AK

EMDB-27233: 
Tomogram of Flublok vaccine hemagglutinin starfish complexes
Method: electron tomography / : Gallagher JR, Audray AK

EMDB-26653: 
Native Lassa glycoprotein in complex with neutralizing antibodies 8.9F and 37.2D
Method: single particle / : Li H, Saphire EO

EMDB-26655: 
Native Lassa glycoprotein in complex with neutralizing antibodies 12.1F and 37.2D
Method: single particle / : Li H, Saphire EO

EMDB-22913: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 1)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22914: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 2)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22915: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)
Method: single particle / : Barnes CO, Bjorkman PJ

EMDB-22916: 
Structure of the SARS-CoV-2 S 6P trimer in complex with the ACE2 protein decoy, CTC-445.2 (State 4)
Method: single particle / : Barnes CO, Bjorkman PJ
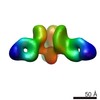
EMDB-7908: 
VIC170 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7909: 
VIC169 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
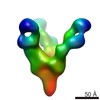
EMDB-7910: 
VIC167 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
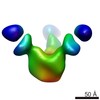
EMDB-7911: 
VIC166 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
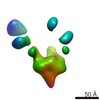
EMDB-7912: 
VIC165 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7914: 
VIC1164 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
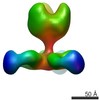
EMDB-7915: 
VIC163 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7926: 
VIC82 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
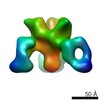
EMDB-7927: 
VIC43 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7928: 
VIC32 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7929: 
VIC26 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7930: 
VIC16 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7931: 
VIC15 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7932: 
VIC12 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
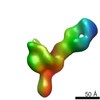
EMDB-7933: 
VIC11 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7922: 
VIC133 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7923: 
VIC101 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7924: 
VIC94 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7925: 
VIC91 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7934: 
VIC130 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
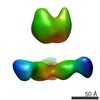
EMDB-7916: 
VIC161 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7917: 
VIC160 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
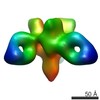
EMDB-7918: 
VIC158 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
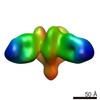
EMDB-7919: 
VIC157 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-7920: 
VIC142 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB
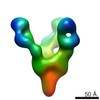
EMDB-7921: 
VIC139 Fab in complex with Ebola virus GP
Method: single particle / : Turner H, Murin CD, Pallesen J, Ward AB

EMDB-6586: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies
Method: single particle / : Borndoldt ZA, Turner HL, Murin CD, Li W, Sok D, Sounders C, Piper AE, Fusco ML, Pommert K, Klempner M, Reimann KA, Golf A, Wittrup DK, Gerngross TU, Krauland E, Glass PJ, Burton DR, Saphire EO, Ward AB, Walker LM

EMDB-6587: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies
Method: single particle / : Borndoldt ZA, Turner HL, Murin CD, Li W, Sok D, Sounders C, Piper AE, Fusco ML, Pommert K, Klempner M, Reimann KA, Golf A, Wittrup DK, Gerngross TU, Krauland E, Glass PJ, Burton DR, Saphire EO, Ward AB, Walker LM
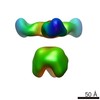
EMDB-6588: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies
Method: single particle / : Borndoldt ZA, Turner HL, Murin CD, Li W, Sok D, Sounders C, Piper AE, Fusco ML, Pommert K, Klempner M, Reimann KA, Golf A, Wittrup DK, Gerngross TU, Krauland E, Glass PJ, Burton DR, Saphire EO, Ward AB, Walker LM
EMDB-6589: 
Negative stain EM of Ebola virus GP bound to human survivor antibodies
Method: single particle / : Borndoldt ZA, Turner HL, Murin CD, Li W, Sok D, Sounders C, Piper AE, Fusco ML, Pommert K, Klempner M, Reimann KA, Golf A, Wittrup DK, Gerngross TU, Krauland E, Glass PJ, Burton DR, Saphire EO, Ward AB, Walker LM

EMDB-2801: 
Negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri
Method: single particle / : Cheung M, Shen D, Makino F, Kato T, Roehrich D, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ

EMDB-2802: 
Negative stain electron microscopy reconstruction of the tip complex from the type III secretion system of Shigella flexneri lacking IpaB
Method: single particle / : Cheung M, Shen D, Makino F, Kato T, Roehrich D, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ

EMDB-2803: 
Negative stain electron microscopy reconstruction of the tip complex and needle from the type III secretion system of Shigella flexneri with MxiH mutation P44A
Method: single particle / : Cheung M, Shen D, Makino F, Kato T, Roehrich D, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ

EMDB-2805: 
Refined negative stain electron microscopy reconstruction of the tip complex from the type III secretion system of Shigella flexneri with MxiH mutation Q51A
Method: single particle / : Cheung M, Shen D, Makino F, Kato T, Roehrich D, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ

EMDB-2806: 
Negative stain electron microscopy reconstruction of the tip complex from the type III secretion system of Shigella flexneri with MxiH mutations P44A+Q51A
Method: single particle / : Cheung M, Shen D, Makino F, Kato T, Roehrich D, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ

EMDB-2804: 
Negative stain electron microscopy reconstruction of the tip complex from the type III secretion system of Shigella flexneri with MxiH mutation Q51A
Method: single particle / : Cheung M, Shen D, Makino F, Kato T, Roehrich D, Martinez-Argudo I, Walker ML, Murillo I, Liu X, Pain M, Brown J, Frazer G, Mantell J, Mina P, Todd T, Sessions RB, Namba K, Blocker AJ

EMDB-2155: 
Cryo-EM structure of axonemal dynein-c prepared in the absence of nucleotide
Method: single particle / : Roberts AJ, Malkova B, Walker ML, Sakakibara H, Numata N, Kon T, Ohkura R, Edwards TA, Knight PJ, Sutoh K, Oiwa K, Burgess SA

EMDB-2156: 
Cryo-EM structure of axonemal dynein-c in the ADP.Vi state
Method: single particle / : Roberts AJ, Malkova B, Walker ML, Sakakibara H, Numata N, Kon T, Ohkura R, Edwards TA, Knight PJ, Sutoh K, Oiwa K, Burgess SA
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
