-Search query
-Search result
Showing 1 - 50 of 178 items for (author: tong & ki)

EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD

EMDB-29677: 
Structure of the methylosome-Lsm10/11 complex
Method: single particle / : Lin M, Paige A, Tong L

PDB-8g1u: 
Structure of the methylosome-Lsm10/11 complex
Method: single particle / : Lin M, Paige A, Tong L

EMDB-29725: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

EMDB-29731: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g4m: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

PDB-8g4t: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD
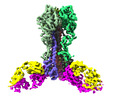
EMDB-27139: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

PDB-8d21: 
Cryo-EM structure of the VRC321 clinical trial, vaccine-elicited, human antibody 1B06 in complex with a stabilized NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-33241: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ

PDB-7xk2: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ

EMDB-26044: 
H10ssF: ferritin-based nanoparticle displaying H10 hemagglutinin stabilized stem epitopes
Method: single particle / : Gallagher JR, Harris AK

EMDB-25794: 
Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-25797: 
Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-25806: 
Locally refined region of SARS-CoV-2 spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

EMDB-25807: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD
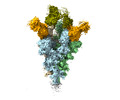
EMDB-25808: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibodies A19-46.1 and B1-182.1
Method: single particle / : Zhou T, Kwong PD

EMDB-26256: 
Local refinement of cryo-EM structure of the interface of the Omicron RBD in complex with antibodies B-182.1 and A19-46.1
Method: single particle / : Zhou T, kwong PD

PDB-7tb8: 
Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tbf: 
Locally refined region of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tc9: 
Locally refined region of SARS-CoV-2 spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

PDB-7tca: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1
Method: single particle / : Zhou T, Kwong PD

PDB-7tcc: 
Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibodies A19-46.1 and B1-182.1
Method: single particle / : Zhou T, Kwong PD

PDB-7u0d: 
Local refinement of cryo-EM structure of the interface of the Omicron RBD in complex with antibodies B-182.1 and A19-46.1
Method: single particle / : Zhou T, kwong PD

EMDB-24060: 
Structure of the SARS-CoV-2 Spike trimer with all RBDs down in complex with the Fab fragment of human neutralizing antibody clone 6
Method: single particle / : Hu Y, Xiong Y

EMDB-24061: 
Structure of the SARS-CoV-2 Spike trimer with two RBDs down in complex with the Fab fragment of human neutralizing antibody clone 6
Method: single particle / : Hu Y, Xiong Y

EMDB-24062: 
Structure of the SARS-CoV-2 Spike trimer with one RBD down in complex with the Fab fragment of human neutralizing antibody clone 6
Method: single particle / : Hu Y, Xiong Y

EMDB-24063: 
Structure of the SARS-CoV-2 Spike trimer with one RBD down in complex with the Fab fragment of human neutralizing antibody clone 2
Method: single particle / : Hu Y, Xiong Y

EMDB-24064: 
Structure of the SARS-CoV-2 Spike trimer with three RBDs up in complex with the Fab fragment of human neutralizing antibody clone 2
Method: single particle / : Hu Y, Xiong Y

PDB-7mw2: 
Structure of the SARS-CoV-2 Spike trimer with all RBDs down in complex with the Fab fragment of human neutralizing antibody clone 6
Method: single particle / : Hu Y, Xiong Y

PDB-7mw3: 
Structure of the SARS-CoV-2 Spike trimer with two RBDs down in complex with the Fab fragment of human neutralizing antibody clone 6
Method: single particle / : Hu Y, Xiong Y

PDB-7mw4: 
Structure of the SARS-CoV-2 Spike trimer with one RBD down in complex with the Fab fragment of human neutralizing antibody clone 6
Method: single particle / : Hu Y, Xiong Y

PDB-7mw5: 
Structure of the SARS-CoV-2 Spike trimer with one RBD down in complex with the Fab fragment of human neutralizing antibody clone 2
Method: single particle / : Hu Y, Xiong Y

PDB-7mw6: 
Structure of the SARS-CoV-2 Spike trimer with three RBDs up in complex with the Fab fragment of human neutralizing antibody clone 2
Method: single particle / : Hu Y, Xiong Y

EMDB-23480: 
Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

PDB-7lpn: 
Cryo-EM structure of llama J3 VHH antibody in complex with HIV-1 Env BG505 DS-SOSIP.664
Method: single particle / : Gorman J, Kwong PD

EMDB-25792: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

PDB-7tb4: 
Cryo-EM structure of the spike of SARS-CoV-2 Omicron variant of concern
Method: single particle / : Zhou T, Tsybovsky T, Kwong PD

EMDB-30956: 
Cryo-EM structure of hDisp1NNN-3C
Method: single particle / : Li W, Wang L, Gong X

EMDB-30957: 
Cryo-EM structure of hDisp1NNN-3C-Cleavage
Method: single particle / : Li W, Wang L, Gong X

EMDB-30958: 
Cryo-EM structure of hDisp1NNN-ShhN
Method: single particle / : Li W, Wang L, Gong X

PDB-7e2h: 
Cryo-EM structure of hDisp1NNN-3C-Cleavage
Method: single particle / : Li W, Wang L, Gong X

EMDB-25105: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

PDB-7sg4: 
Structure of SARS-CoV S protein in complex with Receptor Binding Domain antibody DH1047
Method: single particle / : Gobeil S, Acharya P

EMDB-23098: 
Cryo-EM structure of the VRC316 clinical trial, vaccine-elicited, human antibody 316-310-1B11 in complex with an H2 CAN05 HA trimer
Method: single particle / : Gorman J, Kwong PD

EMDB-23816: 
Cryo-EM structure of the VRC310 clinical trial, vaccine-elicited, human antibody 310-030-1D06 Fab in complex with an H1 NC99 HA trimer
Method: single particle / : Gorman J, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
