-Search query
-Search result
Showing all 24 items for (author: thomson & e)

EMDB-13986: 
Fibrillin-1 microfibril arm-region
Method: single particle / : Godwin ARF, Thomson J, Holmes DF, Adamo CS, Sengle G, Sherratt MJ, Roseman AM, Baldock C

EMDB-13984: 
Structure of Fibrillin Microfibrils from Cryo-Electron Microscopy reveals the site of latent TGFbeta binding and structural perturbations in disease-linked mutations
Method: single particle / : Godwin ARF, Thomson J, Holmes DF, Adamo CS, Sengle G, Sherratt MJ, Roseman AM, Baldock C

EMDB-27178: 
Structure of Cas12a2 binary complex
Method: single particle / : Bravo JPK, Taylor DW
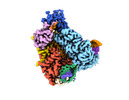
EMDB-27179: 
Cas12a2 quaternary complex
Method: single particle / : Bravo JPK, Taylor DW
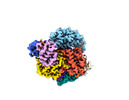
EMDB-27180: 
Structure of Cas12a2 ternary complex
Method: single particle / : Bravo JPK, Taylor DW

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhb: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhc: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-22078: 
Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis
Method: single particle / : Herrera NG, Morano NC, Celikgil A, Georgiev GI, Malonis R, Lee JH, Tong K, Vergnolle O, Massimi A, Yen LY, Noble AJ, Kopylov M, Bonanno JB, Garrett-Thompson SC, Hayes DB, Brenowitz M, Garforth SJ, Eng ET, Lai JR, Almo SC

PDB-6x6p: 
Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis
Method: single particle / : Herrera NG, Morano NC, Celikgil A, Georgiev GI, Malonis R, Lee JH, Tong K, Vergnolle O, Massimi A, Yen LY, Noble AJ, Kopylov M, Bonanno JB, Garrett-Thompson SC, Hayes DB, Brenowitz M, Garforth SJ, Eng ET, Lai JR, Almo SC

EMDB-3745: 
Cryo-EM structure of the foot-containing 80S ribosome
Method: single particle / : Sarkar A, Thoms M, Barrio-Garcia C, Thomson E, Flemming D, Beckmann R, Hurt E

EMDB-3886: 
Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae
Method: single particle / : Heuer A, Thomson E, Schmidt C, Berninghausen O, Becker T, Hurt E, Beckmann R

PDB-6eml: 
Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae
Method: single particle / : Heuer A, Thomson E, Schmidt C, Berninghausen O, Becker T, Hurt E, Beckmann R

EMDB-2110: 
Negative structure of open and closed conformation of crm1
Method: single particle / : Monecke T, Haselbach D, Neumann P, Thomson E, Hurt E, Stark H, Dickmanns A, Ficner R

EMDB-2111: 
Negative structure of closed conformation of crm1
Method: single particle / : Monecke T, Haselbach D, Neumann P, Thomson E, Hurt E, Stark H, Dickmanns A, Ficner R

EMDB-1995: 
Electron density map of a composite coiled-coil fibril comprising multiple self-assembling fibre peptides.
Method: electron crystallography / : Sharp TH, Bruning M, Mantell J, Sessions RB, Thomson AR, Zaccai NR, Brady RL, Verkade P, Woolfson DN
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
