-Search query
-Search result
Showing all 31 items for (author: thomas & gj)

EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS
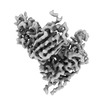
EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS
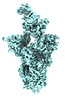
EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tm1: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tma: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS
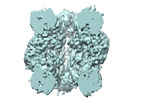
EMDB-27654: 
A subtomogram average of H. neapolitanus Rubisco within alpha-carboxysomes
Method: subtomogram averaging / : Metskas LA, Blikstad C, Laughlin T, Savage DF, Jensen GJ

EMDB-25824: 
aRML prion fibril
Method: single particle / : Hoyt F, Standke HG, Artikis E, Caughey B, Kraus A

EMDB-11928: 
SctV (SsaV) cytoplasmic domain
Method: single particle / : Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T, Yu XJ, Grabe G, Holden D, Nans A, Rosenthal P, Rouse S, Beeby M

EMDB-22975: 
Lethocerus Myosin II complete coiled-coil domain resolved in its native environment
Method: single particle / : Rahmani H, Hu Z, Daneshparvar N, Taylor D, Taylor KA

PDB-7kog: 
Lethocerus Myosin II complete coiled-coil domain resolved in its native environment
Method: single particle / : Rahmani H, Hu Z, Daneshparvar N, Taylor D, Taylor KA

PDB-7beq: 
MicroED structure of the MyD88 TIR domain higher-order assembly
Method: electron crystallography / : Clabbers MTB, Holmes S, Muusse TW, Vajjhala P, Thygesen SJ, Malde AK, Hunter DJB, Croll TI, Nanson JD, Rahaman MH, Aquila A, Hunter MS, Liang M, Yoon CH, Zhao J, Zatsepin NA, Abbey B, Sierecki E, Gambin Y, Darmanin C, Kobe B, Xu H, Ve T

EMDB-20926: 
Clostridium difficile binary toxin translocase CDTb in asymmetric tetradecamer conformation
Method: single particle / : Xu X, Pozharski E

EMDB-20927: 
Clostridium difficile binary toxin translocase CDTb tetradecamer in symmetric conformation
Method: single particle / : Xu X, Pozharski E

PDB-6uwr: 
Clostridium difficile binary toxin translocase CDTb in asymmetric tetradecamer conformation
Method: single particle / : Xu X, Pozharski E, des Georges A

PDB-6uwt: 
Clostridium difficile binary toxin translocase CDTb tetradecamer in symmetric conformation
Method: single particle / : Xu X, Pozharski E, des Georges A

EMDB-0436: 
Near-atomic structure of icosahedrally averaged PBCV-1 capsid
Method: single particle / : Fang Q, Rossmann MG

PDB-6ncl: 
Near-atomic structure of icosahedrally averaged PBCV-1 capsid
Method: single particle / : Fang Q, Rossmann MG

EMDB-9004: 
Cryo-EM structure of C. elegans GDP-microtubule
Method: single particle / : Chaaban S, Jariwala S

EMDB-7459: 
Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Carragher B, Potter CS, Kwong PD

EMDB-7460: 
Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Carragher B, Potter CS, Kwong PD

PDB-6cde: 
Cryo-EM structure at 3.8 A resolution of vaccine-elicited antibody vFP20.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Xu K, Liu K, Carragher B, Potter CS, Kwong PD

PDB-6cdi: 
Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122
Method: single particle / : Acharya P, Xu K, Liu K, Carragher B, Potter CS, Kwong PD

EMDB-8420: 
Cryo-EM structure of BG505 DS-SOSIP HIV-1 Env trimer in complex with vaccine elicited, fusion peptide-directed antibody vFP1.01
Method: single particle / : Acharya P, Kwong PD, Potter CS, Carragher B

EMDB-8421: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01
Method: single particle / : Acharya P, Kwong PD, Potter CS, Carragher B

EMDB-8422: 
Cryo-EM structure of an asymmetric complex of BG505 DS-SOSIP HIV-1 Env trimer with vaccine elicited, fusion peptide-directed antibody vFP5.01
Method: single particle / : Acharya P, Kwong PD, Potter CS, Carragher B

EMDB-2170: 
Thermococcus kodakaraensis 70S ribosome
Method: single particle / : Armache JP, Anger AM, Marquez V, Frankenberg S, Froehlich T, Villa E, Berninghausen O, Thomm M, Arnold GJ, Beckmann R, Wilson DN

EMDB-2171: 
Staphylothermus marinus 50S ribosomal subunit
Method: single particle / : Armache JP, Anger AM, Marquez V, Frankenberg S, Froehlich T, Villa E, Berninghausen O, Thomm M, Arnold GJ, Beckmann R, Wilson DN

EMDB-2172: 
Methanococcus igneus 70S ribosome
Method: single particle / : Armache JP, Anger AM, Marquez V, Frankenberg S, Froehlich T, Villa E, Berninghausen O, Thomm M, Arnold GJ, Beckmann R, Wilson DN

EMDB-1796: 
Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins
Method: single particle / : Marquez V, Froehlich T, Armache JP, Sohmen D, Doenhoefer A, Berninghausen O, Thomm M, Beckmann R, Arnold GJ, Wilson DN

EMDB-1797: 
Proteomic characterization of archaeal ribosomes reveals the presence of novel archaeal-specific ribosomal proteins. 50S ribosomal subunit of Sulfolobus acidocaldarius
Method: single particle / : Marquez V, Froehlich T, Armache JP, Sohmen D, Doenhoefer A, Berninghausen O, Thomm M, Beckmann R, Arnold GJ, Wilson DN
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
