-Search query
-Search result
Showing 1 - 50 of 54 items for (author: terry & c)

EMDB-29637: 
Dehosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR)
Method: single particle / : Levring J, Terry DS, Kilic Z, Fitzgerald GA, Blanchard SC, Chen J

PDB-8fzq: 
Dehosphorylated, ATP-bound human cystic fibrosis transmembrane conductance regulator (CFTR)
Method: single particle / : Levring J, Terry DS, Kilic Z, Fitzgerald GA, Blanchard SC, Chen J
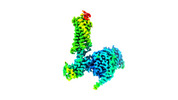
EMDB-29301: 
Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator
Method: single particle / : Krumm BE, Diberto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

EMDB-29302: 
CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL
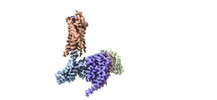
EMDB-29303: 
CryoEM structure of Go-coupled NTSR1
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

PDB-8fmz: 
Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator
Method: single particle / : Krumm BE, Diberto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

PDB-8fn0: 
CryoEM structure of Go-coupled NTSR1 with a biased allosteric modulator
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

PDB-8fn1: 
CryoEM structure of Go-coupled NTSR1
Method: single particle / : Krumm BE, DiBerto JF, Olsen RHJ, Kang H, Slocum ST, Zhang S, Strachan RT, Fay JF, Roth BL

EMDB-33241: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ

PDB-7xk2: 
Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex
Method: single particle / : Yang Y, Kang HJ, Gao RG, Wang JJ, Han GW, DiBerto JF, Wu LJ, Tong JH, Qu L, Wu YR, Pileski R, Li XM, Zhang XC, Zhao SW, Kenakin T, Wang Q, Stevens RC, Peng W, Roth BL, Rao ZH, Liu ZJ
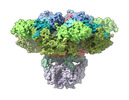
EMDB-27982: 
Structure of the full-length IP3R1 channel determined at high Ca2+
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II
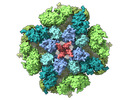
EMDB-27983: 
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II

PDB-8eaq: 
Structure of the full-length IP3R1 channel determined at high Ca2+
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II

PDB-8ear: 
Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP
Method: single particle / : Fan G, Baker MR, Terry LE, Arige V, Chen M, Seryshev AB, Baker ML, Ludtke SJ, Yule DI, Serysheva II
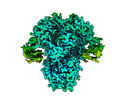
EMDB-26727: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
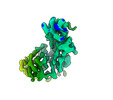
EMDB-26729: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
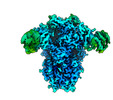
EMDB-26730: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)
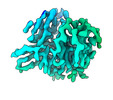
EMDB-26731: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7us6: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7us9: 
CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7usa: 
Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-7usb: 
CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0)
Method: single particle / : Tortorici MA, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-23494: 
Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

EMDB-23495: 
Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

EMDB-23496: 
Cryo-EM of the SLFN12-PDE3A complex: SLFN12 body refinement
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

PDB-7lrc: 
Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

PDB-7lrd: 
Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

PDB-7lre: 
Cryo-EM of the SLFN12-PDE3A complex: SLFN12 body refinement
Method: single particle / : Fuller JR, Garvie CW, Lemke CT

EMDB-20121: 
Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA
Method: single particle / : Hoffer ED, Mehrani A, Keiler KC, Stagg SM, Dunham CM

PDB-6om6: 
Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA
Method: single particle / : Hoffer ED, Mehrani A, Keiler KC, Stagg SM, Dunham CM

EMDB-21411: 
HIV envelope glycoprotein bound with soluble CD4 (D1-D2) and antibody 17b on AT-2 treated BaL strain virus
Method: subtomogram averaging / : Jun L, Ze L, Li W

EMDB-21412: 
Ligand-free HIV envelope glycoprotein on AT-2 treated BaL strain virus
Method: subtomogram averaging / : Jun L, Ze L, Li W

EMDB-21413: 
HIV envelope glycoprotein bound with antibodies 10-1074 and 3BNC117 on AT-2 treated BaL strain virus
Method: subtomogram averaging / : Jun L, Ze L, Li W

EMDB-0173: 
Complex of foot-and-mouth-disease virus (type O1 M) with the Fab of antibody D9. The virus is at 3.5 Ang resolution, the Fab is flexibly attached and at much lower resolution.
Method: single particle / : Shimmon G, Kotecha A, Ren J, Asfor AS, Newman J, Berryman S, Cottam EM, Gold S, Tuthill TJ, King DP, Brocchi E, King AMQ, Owens R, Fry EE, Stuart DI, Burman A, Jackson T

PDB-5ler: 
Structure of the bacterial sex F pilus (13.2 Angstrom rise)
Method: helical / : Costa TRD, Ilangovan I, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G

PDB-5lfb: 
Structure of the bacterial sex F pilus (12.5 Angstrom rise)
Method: helical / : Costa TRD, Ilangovan I, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G

PDB-5leg: 
Structure of the bacterial sex F pilus (pED208)
Method: helical / : Costa TRD, Ilangovan I, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G

EMDB-4042: 
Cryo-EM structure of the bacterial sex F pilus (pED208)
Method: helical / : Costa TRD, Ilangovan I, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G

EMDB-4044: 
Cryo-EM structure of the bacterial sex F pilus (13.2 Angstrom rise)
Method: helical / : Costa TRD, Ilangovan I, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G

EMDB-4046: 
Cryo-EM structure of the bacterial sex F pilus (12.5 Angstrom rise)
Method: helical / : Costa TRD, Ilangovan I, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G

EMDB-4092: 
Tomographic reconstruction of ex vivo mammalian prions
Method: electron tomography / : Terry C, Wenborn A, Gros N, Sells J, Joiner S, Hosszu LLP, Tattum MH, Panico S, Clare DK, Collinge J, Saibil HR, Wadsworth JDF

EMDB-3129: 
Structure-based energetics of protein interfaces guide FMDV vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, Perez-Martin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

EMDB-3130: 
Structure-based energetics of protein interfaces guide FMDV vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, Perez-Martin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

PDB-5ac9: 
Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, PerezMartin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

PDB-5aca: 
Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, Perez-Martin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

EMDB-2433: 
Amyloid-beta nanotube
Method: single particle / : Nicoll AJ, Panico S, Freir DB, Wright D, Terry C, Risse E, Herron CE, O'Malley T, Wadsworth JD, Farrow MA, Walsh DM, Saibil HR, Collinge J

EMDB-5041: 
Ribosome structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5042: 
Lumazine synthase structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5043: 
GroEL structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM

EMDB-5044: 
RNA polymerase structure : Structural survey of large protein complexes in Desulfovibrio vulgaris Hildenborough (DvH)
Method: single particle / : Han BG, Dong M, Liu H, Camp L, Geller J, Singer M, Hazen TC, Choi M, Witkowska HE, Ball DA, Typke D, Downing KH, Shatsky M, Brenner SE, Chandonia JM, Biggin MD, Glaeser RM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
