-Search query
-Search result
Showing 1 - 50 of 70 items for (author: taylor & ik)

EMDB-41725: 
Structural and biochemical rationale for Beta variant protein booster vaccine broad cross-neutralization of SARS-CoV-2
Method: single particle / : Bruch EM, Rak A

EMDB-41727: 
Structural and biochemical rationale for Beta variant protein booster vaccine broad cross-neutralization of SARS-CoV-2
Method: single particle / : Bruch EM, Rak A

PDB-8tyl: 
Structural and biochemical rationale for Beta variant protein booster vaccine broad cross-neutralization of SARS-CoV-2
Method: single particle / : Bruch EM, Rak A

PDB-8tyo: 
Structural and biochemical rationale for Beta variant protein booster vaccine broad cross-neutralization of SARS-CoV-2
Method: single particle / : Bruch EM, Rak A

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
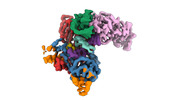
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
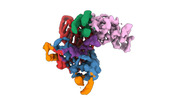
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-26735: 
Hantavirus ANDV Gn(H) protein in complex with 2 Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE
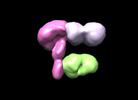
EMDB-26736: 
Hantavirus MAPV Gn(H)/Gc protein in complex with 2 Fabs SNV-24 and SNV-53
Method: single particle / : Binshtein E, Crowe JE

EMDB-27318: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE

PDB-8dbz: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE

EMDB-26322: 
MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution
Method: single particle / : Shan Z, Pye VE, Cherepanov P, Lyumkis D

PDB-7u32: 
MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution
Method: single particle / : Shan Z, Pye VE, Cherepanov P, Lyumkis D

EMDB-14860: 
Cryo-EM structure of the MVV CSC intasome at 4.5A resolution
Method: single particle / : Ballandras-Colas A, Maskell D, Pye VE, Locke J, Swuec S, Kotecha A, Costa A, Cherepanov P

PDB-7zpp: 
Cryo-EM structure of the MVV CSC intasome at 4.5A resolution
Method: single particle / : Ballandras-Colas A, Maskell D, Pye VE, Locke J, Swuec S, Kotecha A, Costa A, Cherepanov P

EMDB-25026: 
Anaphase Promoting Complex delta APC7
Method: single particle / : Ferguson CJ, Brown NG, Prabu JR, Watson ER, Schulman BA, Bonni A

EMDB-25027: 
Anaphase Promoting Complex delta APC7 + UBE2C,substrate,UbV,CDH1
Method: single particle / : Ferguson CJ, Brown NG, Prabu JR, Watson ER, Schulman BA, Bonni A

EMDB-24408: 
The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25
Method: single particle / : Baker AT, Boyd RJ, Sarkar D, Vermaas JV, Williams D, Singharoy A

PDB-7rd1: 
The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25
Method: single particle / : Baker AT, Boyd RJ, Sarkar D, Vermaas JV, Williams D, Singharoy A

EMDB-30707: 
Hemagglutinin Influenza A virus (A/Okuda/1957(H2N2) bound with a neutralizing antibody
Method: single particle / : Shirouzu M

EMDB-24404: 
Covalent inhibition of hAChE by organophosphates causes homodimer dissociation through long-range allosteric effects
Method: single particle / : Blumenthal DK, Cheng X, Fager M, Ho KY, Rohrer J, Gerlits O, Juneja P, Kovalevsky A, Radic Z

EMDB-23614: 
SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-1)
Method: single particle / : Kern DM, Gerber EE, Brohawn SG

EMDB-23616: 
SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-2)
Method: single particle / : Kern DM, Gerber EE, Brohawn SG

PDB-7m17: 
SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-1)
Method: single particle / : Kern DM, Gerber EE, Brohawn SG

PDB-7m19: 
SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-2)
Method: single particle / : Kern DM, Gerber EE, Brohawn SG

EMDB-22738: 
CryoEM reconstruction of SARS-CoV-2 receptor binding domain in complex with the Fab fragment of neutralizing antibody 46
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-22739: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 80.
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-22740: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 298
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP

EMDB-22741: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with the Fab fragment of neutralizing antibody 324
Method: single particle / : Kucharska I, Tan YZ, Benlekbir S, Rubinstein JL, Julien JP
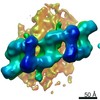
EMDB-20825: 
In situ structure of LRRK2(I2020T)-Microtubule: Microtubule_bound_LRRK2(I2020T)_Tight Mask_A
Method: subtomogram averaging / : Boehning J, Buschauer R, Watanabe R, Villa E

EMDB-20826: 
In situ structure of Parkinson's disease-linked Microtubule-Bound human LRRK2(I2020T) MASK B
Method: subtomogram averaging / : Boehning J, Buschauer R, Watanabe R, Villa E

EMDB-20827: 
Cryo-electron tomogram of FIB-milled HEK293 cells expressing pathogenic LRRK2(I2020T)mutant
Method: electron tomography / : Watanabe R, Villa E

EMDB-20828: 
Cryo-electron tomogram of FIB-milled HEK293 cells treated with Taxol
Method: electron tomography / : Watanabe R, Villa E

PDB-6xr4: 
Integrative in situ structure of Parkinsons disease-linked human LRRK2
Method: subtomogram averaging / : Villa E, Lasker K, Audagnotto M

EMDB-0510: 
Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP)
Method: single particle / : Huang W, Taylor D

PDB-6nts: 
Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP)
Method: single particle / : Huang W, Taylor D

EMDB-7559: 
Bacteriophage A511 baseplate in post-host attachment state (urea-induced)
Method: single particle / : Guerrero-Ferreira R

EMDB-7560: 
Bacteriophage A511 baseplate in pre-host attachment state
Method: single particle / : Guerrero-Ferreira R

EMDB-7561: 
Bacteriophage A511 baseplate in post-host attachment state
Method: single particle / : Guerrero-Ferreira R

EMDB-3792: 
Electron cryotomogram of Vibrio cholerae O395 N1
Method: electron tomography / : Chang YW, Kjaer A, Ortega DR, Kovacikova G, Sutherland JA, Rettberg LA, Taylor RK, Jensen GJ

EMDB-8492: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8493: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8494: 
Subtomogram average of the piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8495: 
Subtomogram average of the piliated toxin-coregulated pilus machine in wild-type Vibrio cholerae cells (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8496: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpS knockout (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8497: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpS knockout (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8498: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpB knockout (aligning OM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ

EMDB-8499: 
Subtomogram average of the non-piliated toxin-coregulated pilus machine in Vibrio cholerae cells with tcpB knockout (aligning IM-parts)
Method: subtomogram averaging / : Chang YW, Kjaer A, Jensen GJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
