-Search query
-Search result
Showing 1 - 50 of 170 items for (author: tan & lc)
EMDB-41299: 
Structural basis of peptidoglycan synthesis by E. coli RodA-PBP2 complex
Method: single particle / : Nygaard R, Mancia F
EMDB-41303: 
Transmembrane map
Method: single particle / : Nygaard R, Mancia F
EMDB-41304: 
Periplasmic map
Method: single particle / : Nygaard R, Mancia F

EMDB-16677: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 1)
Method: helical / : Arseni D, Ryskeldi-Falcon B

EMDB-16681: 
Structure of TDP-43 amyloid filaments from type A FTLD-TDP (individual 2, variant 2)
Method: helical / : Arseni D, Ryskeldi-Falcon B

EMDB-16682: 
Structure of TDP-43 amyloid filaments from type A FTLD-TDP (individual 3, variant 1)
Method: helical / : Arseni D, Ryskeldi-Falcon B

EMDB-16628: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 1)
Method: helical / : Arseni D, Ryskeldi-Falcon B

EMDB-16642: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 2)
Method: helical / : Arseni D, Ryskeldi-Falcon B

EMDB-16643: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 3)
Method: helical / : Arseni D, Ryskeldi-Falcon B

PDB-8cg3: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 1)
Method: helical / : Arseni D, Ryskeldi-Falcon B

PDB-8cgg: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 2)
Method: helical / : Arseni D, Ryskeldi-Falcon B

PDB-8cgh: 
Structure of TDP-43 amyloid filament from type A FTLD-TDP (variant 3)
Method: helical / : Arseni D, Ryskeldi-Falcon B

EMDB-29686: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI19 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29704: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 3 FNI9 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29705: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI9 Fab molecules
Method: single particle / : Dang H, Snell G

EMDB-29706: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI9 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29707: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI9 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29708: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 3 FNI17 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29709: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 with S245N S247T mutations in complex with one FNI17 Fab molecule
Method: single particle / : Dang HV, Snell G

EMDB-29710: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI19 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29711: 
Neuraminidase of B/Massachusetts/02/2012 (Yamagata) in complex with 4 FNI17 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-29712: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI19 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g30: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI19 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g3m: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 3 FNI9 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g3n: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI9 Fab molecules
Method: single particle / : Dang H, Snell G

PDB-8g3o: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI9 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g3p: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI9 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g3q: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 3 FNI17 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g3r: 
N2 neuraminidase of A/Tanzania/205/2010 H3N2 with S245N S247T mutations in complex with one FNI17 Fab molecule
Method: single particle / : Dang HV, Snell G

PDB-8g3v: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI19 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g3z: 
Neuraminidase of B/Massachusetts/02/2012 (Yamagata) in complex with 4 FNI17 Fab molecules
Method: single particle / : Dang HV, Snell G

PDB-8g40: 
N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI19 Fab molecules
Method: single particle / : Dang HV, Snell G

EMDB-16703: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16704: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16705: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16706: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16707: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Nup153 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16708: 
HIV-1 mature capsid hexamer next to pentamer (type I) from CA-IP6 CLPs bound to Nup153 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16709: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to CPSF6 peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16710: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to CPSF6 peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16711: 
HIV-1 mature capsid hexamer from CA-IP6 CLPs, bound to Sec24C peptide.
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-16712: 
HIV-1 mature capsid pentamer from CA-IP6 CLPs bound to Sec24C peptide
Method: single particle / : Stacey JCV, Briggs JAG

EMDB-25567: 
Cryo-EM structure of OmpK36-TraN mating pair stabilization proteins from carbapenem-resistant Klebsiella pneumoniae
Method: single particle / : Beltran LC, Seddon C, Beis K, Frankel G, Egelman EH

EMDB-11953: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-11954: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14810: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14811: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement
Method: single particle / : Hurdiss DL, Drulyte I
EMDB-26054: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
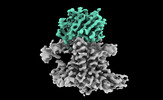
EMDB-26057: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

PDB-7tpg: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
