-Search query
-Search result
Showing 1 - 50 of 149 items for (author: steyaert & j)
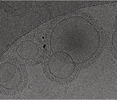
EMDB-16813: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ

EMDB-16814: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ

EMDB-16815: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ

EMDB-16794: 
Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3
Method: single particle / : Kuhm TI, Jakobi AJ

EMDB-41062: 
CryoEM structure of an inward-facing MelBSt at a Na(+)-bound and sugar low-affinity conformation
Method: single particle / : Guan L

PDB-8t60: 
CryoEM structure of an inward-facing MelBSt at a Na(+)-bound and sugar low-affinity conformation
Method: single particle / : Guan L

EMDB-18541: 
Structure of the mu opioid receptor bound to the antagonist nanobody NbE
Method: single particle / : Yu J, Kumar A, Zhang X, Martin C, Raia P, Manglik A, Ballet S, Boland A, Stoeber M

PDB-8qot: 
Structure of the mu opioid receptor bound to the antagonist nanobody NbE
Method: single particle / : Yu J, Kumar A, Zhang X, Martin C, Raia P, Manglik A, Ballet S, Boland A, Stoeber M

EMDB-17537: 
Structure of 5D3-Fab and nanobody(Nb8)-bound ABCG2
Method: single particle / : Irobalieva RN, Manolaridis I, Jackson SM, Ni D, Pardon E, Stahlberg H, Steyaert J, Locher KP

EMDB-17543: 
Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2
Method: single particle / : Irobalieva RN, Manolaridis I, Jackson SM, Ni D, Pardon E, Stahlberg H, Steyaert J, Locher KP

EMDB-17547: 
Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2
Method: single particle / : Irobalieva RN, Manolaridis I, Jackson SM, Ni D, Pardon E, Stahlberg H, Steyaert J, Locher KP

PDB-8p7w: 
Structure of 5D3-Fab and nanobody(Nb8)-bound ABCG2
Method: single particle / : Irobalieva RN, Manolaridis I, Jackson SM, Ni D, Pardon E, Stahlberg H, Steyaert J, Locher KP

PDB-8p8a: 
Structure of 5D3-Fab and nanobody(Nb17)-bound ABCG2
Method: single particle / : Irobalieva RN, Manolaridis I, Jackson SM, Ni D, Pardon E, Stahlberg H, Steyaert J, Locher KP

PDB-8p8j: 
Structure of 5D3-Fab and nanobody(Nb96)-bound ABCG2
Method: single particle / : Irobalieva RN, Manolaridis I, Jackson SM, Ni D, Pardon E, Stahlberg H, Steyaert J, Locher KP

EMDB-29857: 
Molecular mechanism of nucleotide inhibition of human uncoupling protein 1
Method: single particle / : Gogoi P, Jones SA, Ruprecht JJ, King MS, Lee Y, Zogg T, Pardon E, Chand D, Steimle S, Copeman D, Cotrim CA, Steyaert J, Crichton PG, Moiseenkova-Bell V, Kunji ERS

PDB-8g8w: 
Molecular mechanism of nucleotide inhibition of human uncoupling protein 1
Method: single particle / : Gogoi P, Jones SA, Ruprecht JJ, King MS, Lee Y, Zogg T, Pardon E, Chand D, Steimle S, Copeman D, Cotrim CA, Steyaert J, Crichton PG, Moiseenkova-Bell V, Kunji ERS

EMDB-14618: 
Dipeptide and tripeptide Permease C (DtpC)
Method: single particle / : Killer M, Finocchio G, Pardon E, Steyaert J, Loew C

PDB-7zc2: 
Dipeptide and tripeptide Permease C (DtpC)
Method: single particle / : Killer M, Finocchio G, Pardon E, Steyaert J, Loew C

EMDB-13764: 
Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map)
Method: single particle / : Coupland C, Carrique L, Siebold C

PDB-7q1u: 
Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Composite Map)
Method: single particle / : Coupland C, Carrique L, Siebold C

EMDB-13593: 
Structure of thermostabilised human NTCP in complex with nanobody 87
Method: single particle / : Goutam K, Reyes N

EMDB-13596: 
Structure of thermostabilised human NTCP in complex with Megabody 91
Method: single particle / : Goutam K, Reyes N

PDB-7pqg: 
Structure of thermostabilised human NTCP in complex with nanobody 87
Method: single particle / : Goutam K, Reyes N

PDB-7pqq: 
Structure of thermostabilised human NTCP in complex with Megabody 91
Method: single particle / : Goutam K, Reyes N

EMDB-25366: 
Chlorella virus hyaluronan synthase
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25367: 
Chlorella virus hyaluronan synthase inhibited by UDP
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25368: 
Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25369: 
Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-25370: 
Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

PDB-7sp6: 
Chlorella virus hyaluronan synthase
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

PDB-7sp7: 
Chlorella virus hyaluronan synthase inhibited by UDP
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

PDB-7sp8: 
Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

PDB-7sp9: 
Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

PDB-7spa: 
Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state
Method: single particle / : Maloney FP, Kuklewicz J, Zimmer J

EMDB-14578: 
Structure of Hedgehog acyltransferase (HHAT) in complex with megabody 177 bound to non-hydrolysable palmitoyl-CoA (Consensus Map)
Method: single particle / : Coupland C, Carrique L, Siebold C

EMDB-12605: 
Human TRiC complex in closed state with nanobody bound (Consensus Map)
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W, Structural Genomics Consortium (SGC)

EMDB-12606: 
Human TRiC complex in closed state with nanobody Nb18, actin and PhLP2A bound
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W, Structural Genomics Consortium (SGC)

EMDB-12607: 
Human TRiC complex in closed state with nanobody and tubulin bound
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W

EMDB-12608: 
Human TRiC complex in open state with nanobody bound
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W

PDB-7nvl: 
Human TRiC complex in closed state with nanobody bound (Consensus Map)
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W, Structural Genomics Consortium (SGC)

PDB-7nvm: 
Human TRiC complex in closed state with nanobody Nb18, actin and PhLP2A bound
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W, Structural Genomics Consortium (SGC)

PDB-7nvn: 
Human TRiC complex in closed state with nanobody and tubulin bound
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W

PDB-7nvo: 
Human TRiC complex in open state with nanobody bound
Method: single particle / : Kelly JJ, Chi G, Bulawa C, Paavilainen VO, Bountra C, Huiskonen JT, Yue W

EMDB-23812: 
Structure of the apo phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex
Method: single particle / : Burke JE, Dalwadi U, Rathinaswamy MK, Yip CK

EMDB-14048: 
Torpedo muscle-type nicotinic acetylcholine receptor - Resting conformation
Method: single particle / : Zarkadas E, Pebay-Peyroula E, Baenziger J, Nury H

EMDB-14064: 
Torpedo muscle-type nicotinic acetylcholine receptor - nicotine-bound conformation
Method: single particle / : Zarkadas E, Pebay-Peyroula E, Baenziger J, Nury H

EMDB-14065: 
Torpedo muscle-type nicotinic acetylcholine receptor - carbamylcholine-bound conformation
Method: single particle / : Zarkadas E, Pebay-Peyroula E, Baenziger J, Nury H

PDB-7qko: 
Torpedo muscle-type nicotinic acetylcholine receptor - Resting conformation
Method: single particle / : Zarkadas E, Pebay-Peyroula E, Baenziger J, Nury H

PDB-7ql5: 
Torpedo muscle-type nicotinic acetylcholine receptor - nicotine-bound conformation
Method: single particle / : Zarkadas E, Pebay-Peyroula E, Baenziger J, Nury H

PDB-7ql6: 
Torpedo muscle-type nicotinic acetylcholine receptor - carbamylcholine-bound conformation
Method: single particle / : Zarkadas E, Pebay-Peyroula E, Baenziger J, Nury H
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
