-Search query
-Search result
Showing 1 - 50 of 960 items for (author: stein & ar)
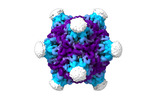
EMDB-19024: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
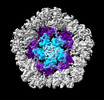
EMDB-19025: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
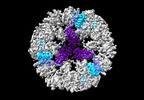
EMDB-19026: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG
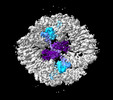
EMDB-19027: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb3: 
Structure of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb4: 
Structure of the five-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb5: 
Structure of the three-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

PDB-8rb7: 
Structure of the two-fold capsomer of the PNMA2 capsid
Method: single particle / : Erlendsson S, Xu J, Shepherd JD, Briggs JAG

EMDB-42371: 
Mouse apoferritin imaged with a square aperture
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42372: 
Mouse apoferritin imaged with a round aperture
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42373: 
Mouse apoferritin imaged with a square aperture with P2 projection lens rotation, reconstructed from 25,000 particles
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42374: 
Mouse apoferritin imaged with a round aperture without P2 projection lens rotation, reconstructed from 25,000 particles
Method: single particle / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-43320: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43321: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43322: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43323: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43324: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43325: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43326: 
Negative Stain EM Reconstructions of SARS-CoV-2 spike proteins mixed with polyclonal antibodies from donor 4.
Method: single particle / : Mannar D, Zhu X, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkk: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkl: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkm: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkn: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vko: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkp: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S
EMDB-42851: 
5x5 tiled montage tomogram of a holey carbon grid with apoferritin imaged with a square electron beam
Method: electron tomography / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-42879: 
3x3 tiled montage tomogram of a yeast lamella imaged with a square electron beam
Method: electron tomography / : Chua EYD, Alink LM, Kopylov M, Johnston J, Einsenstein F, de Marco A

EMDB-18715: 
TDP-43 amyloid fibrils: Morphology-1a
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

EMDB-18716: 
TDP-43 amyloid fibrils: Morphology-1b
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

EMDB-18717: 
TDP-43 amyloid fibrils: Morphology-2
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

PDB-8qx9: 
TDP-43 amyloid fibrils: Morphology-1a
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

PDB-8qxa: 
TDP-43 amyloid fibrils: Morphology-1b
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

PDB-8qxb: 
TDP-43 amyloid fibrils: Morphology-2
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

EMDB-42853: 
Cryo-EM structure of the unliganded hexameric prenyltransferase in bifunctional copalyl diphosphate synthase from Penicillium fellutanum with an open conformation
Method: single particle / : Gaynes MN, Christianson DW

EMDB-42855: 
Cryo-EM structure of the unliganded hexameric prenyltransferase in bifunctional copalyl diphosphate synthase from Penicillium fellutanum with an open conformation, reconstruction in C1
Method: single particle / : Gaynes MN, Christianson DW

PDB-8v0f: 
Cryo-EM structure of the unliganded hexameric prenyltransferase in bifunctional copalyl diphosphate synthase from Penicillium fellutanum with an open conformation
Method: single particle / : Gaynes MN, Christianson DW

EMDB-42510: 
Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway
Method: single particle / : Vasquez S, Drennan CL

EMDB-42511: 
Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway
Method: single particle / : Vasquez S, Drennan CL

EMDB-42512: 
Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway
Method: single particle / : Vasquez S, Drennan C

EMDB-42513: 
Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway
Method: single particle / : Vasquez S, Drennan C

EMDB-42514: 
Structural and biochemical investigations of a HEAT-repeat protein involved in the cytosolic iron-sulfur cluster assembly pathway
Method: single particle / : Vasquez S, Drennan CL

EMDB-41037: 
Cryo-EM structure of immature Zika virus
Method: single particle / : Moustafa IM, Hafenstein SL

EMDB-18941: 
SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules)
Method: single particle / : Guttler T, Aksu M, Gorlich D

EMDB-17736: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17737: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17738: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pke: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkf: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkg: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M
EMDB-41004: 
Heterodimeric ABC transporter BmrCD in the inward-facing conformation bound to ATP: BmrCD_IF-ATP2
Method: single particle / : Tang Q, Mchaourab H
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
