-Search query
-Search result
Showing all 41 items for (author: snyder & m)

EMDB-24479: 
Structure of ACLY-D1026A-substrates
Method: single particle / : Wei X, Marmorstein R

EMDB-24511: 
Structure of ACLY D1026A-substrates-asym-int
Method: single particle / : Wei X, Marmorstein R

EMDB-24577: 
Structure of ACLY D1026A - substrates-asym
Method: single particle / : Wei X, Marmorstein R

EMDB-29668: 
Structure of ACLY-D1026A-products-asym
Method: single particle / : Wei X, Marmorstein R

EMDB-29669: 
Structure of ACLY-D1026A-products
Method: single particle / : Wei X, Marmorstein R

EMDB-29739: 
Structure of ACLY-D1026A-substrates, local refinement of ASH domain
Method: single particle / : Xuepeng W, Ronen M

EMDB-29740: 
Structure of ACLY-D1026A-products, local refinement of ASH domain
Method: single particle / : Wei X, Marmorstein R

PDB-7rig: 
Structure of ACLY-D1026A-substrates
Method: single particle / : Wei X, Marmorstein R

PDB-7rkz: 
Structure of ACLY D1026A-substrates-asym-int
Method: single particle / : Wei X, Marmorstein R

PDB-7rmp: 
Structure of ACLY D1026A - substrates-asym
Method: single particle / : Wei X, Marmorstein R

PDB-8g1e: 
Structure of ACLY-D1026A-products-asym
Method: single particle / : Wei X, Marmorstein R

PDB-8g1f: 
Structure of ACLY-D1026A-products
Method: single particle / : Wei X, Marmorstein R

PDB-8g5c: 
Structure of ACLY-D1026A-substrates, local refinement of ASH domain
Method: single particle / : Xuepeng W, Ronen M

PDB-8g5d: 
Structure of ACLY-D1026A-products, local refinement of ASH domain
Method: single particle / : Wei X, Marmorstein R

EMDB-26547: 
Mediator-PIC Early (Tail A + Upstream DNA & Activator)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26551: 
Mediator-PIC Early (Composite Model)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uik: 
Mediator-PIC Early (Tail A + Upstream DNA & Activator)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uio: 
Mediator-PIC Early (Composite Model)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26542: 
Core Mediator-PICearly (Copy A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26543: 
Mediator-PIC Early (Tail A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26544: 
Mediator-PIC Early (Core B)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26545: 
Mediator-PIC Early (Mediator A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K
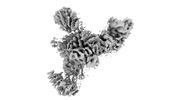
EMDB-26548: 
Mediator-PIC Early (Tail A/B Dimer)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-28023: 
Dimerized Mediator-PIC with early GTFs and TFIIE on a divergent promoter
Method: subtomogram averaging / : Palao III L
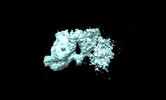
EMDB-28024: 
Dimerized Mediator PIC with early GTFs on a divergent promoter
Method: subtomogram averaging / : Palao III L

PDB-7ui9: 
Core Mediator-PICearly (Copy A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uic: 
Mediator-PIC Early (Tail A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uif: 
Mediator-PIC Early (Core B)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uig: 
Mediator-PIC Early (Mediator A)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

PDB-7uil: 
Mediator-PIC Early (Tail A/B Dimer)
Method: single particle / : Gorbea Colon JJ, Chen SF, Tsai KL, Murakami K

EMDB-26440: 
Ribosome co-immunoprecipitation with Rad6-FLAG showing conventional 40S beak
Method: single particle / : Zhou Y, Bartesaghi A, Silva GM

EMDB-26441: 
Ribosome co-immunoprecipitation with Rad6-FLAG showing extended conformation 40S beak
Method: single particle / : Zhou Y, Bartesaghi A, Silva GM

EMDB-22989: 
Structure of kinase and Central lobes of yeast CKM
Method: single particle / : Li YC, Chao TC

EMDB-22990: 
Structure of the H-lobe of yeast CKM
Method: single particle / : Li YC, Chao TC

EMDB-22991: 
Structure of the yeast CKM
Method: single particle / : Li YC, Chao TC

PDB-7kpv: 
Structure of kinase and Central lobes of yeast CKM
Method: single particle / : Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL

PDB-7kpw: 
Structure of the H-lobe of yeast CKM
Method: single particle / : Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL

PDB-7kpx: 
Structure of the yeast CKM
Method: single particle / : Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Nakanishi K, Chang CE, Murakami K, Garcia BA, Boyer TG, Tsai KL
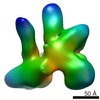
EMDB-1975: 
Functional reconstitution of the 13-subunit human translation initiation factor eIF3.
Method: single particle / : Querol-Audi J, Sun C, Todorovic A, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis C, Hartland A, Gradia S, Fraser CS, Doudna JA, Nogales E, Cate JHD
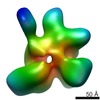
EMDB-1976: 
Functional reconstitution of the 13-subunit human translation initiation factor eIF3.
Method: single particle / : Querol-Audi J, Sun C, Todorovic A, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis C, Hartland A, Gradia S, Fraser CS, Doudna JA, Nogales E, Cate JHD

PDB-1tvk: 
The binding mode of epothilone A on a,b-tubulin by electron crystallography
Method: electron crystallography / : Nettles JH, Li H, Cornett B, Krahn JM, Snyder JP, Downing KH
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
