-Search query
-Search result
Showing 1 - 50 of 210 items for (author: sham & a)

EMDB-19064: 
CryoEM reconstruction of SARS-CoV-2 spike in complex with nanobody tri-TMH (partially open conformation)
Method: single particle / : Rissanen I, Hannula L, Huiskonen JT

EMDB-19068: 
CryoEM reconstruction of SARS-CoV-2 Spike in complex with nanobody tri-TMH (closed conformation)
Method: single particle / : Rissanen I, Hannula L, Huiskonen JT

EMDB-29423: 
Structure of Escherichia coli CedA in complex with transcription initiation complex
Method: single particle / : Liu M, Vassyliev N, Nudler E

PDB-8ftd: 
Structure of Escherichia coli CedA in complex with transcription initiation complex
Method: single particle / : Liu M, Vassyliev N, Nudler E

EMDB-34715: 
Cryo-EM structure of ComA bound to its mature substrate CSP peptide
Method: single particle / : Yu L, Xin X, Min L
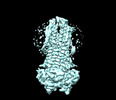
EMDB-36882: 
Cryo-EM structure of nucleotide-bound ComA with ZinC ion
Method: single particle / : Yu L, Xin X, Min L, Feng H

EMDB-36936: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant with Mg2+
Method: single particle / : Yu L, Xin X, Min L

PDB-8hf7: 
Cryo-EM structure of ComA bound to its mature substrate CSP peptide
Method: single particle / : Yu L, Xin X, Min L

PDB-8k4b: 
Cryo-EM structure of nucleotide-bound ComA with ZinC ion
Method: single particle / : Yu L, Xin X, Min L, Feng H

PDB-8k7a: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant with Mg2+
Method: single particle / : Yu L, Xin X, Min L
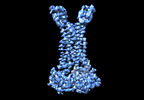
EMDB-35362: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-35363: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M
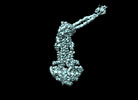
EMDB-35364: 
Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-35437: 
Cryo-EM structure of Mycobacterium tuberculosis ADP bound FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-36304: 
Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8idb: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8idc: 
Cryo-EM structure of Mycobacterium tuberculosis FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8idd: 
Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8igq: 
Cryo-EM structure of Mycobacterium tuberculosis ADP bound FtsEX/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

PDB-8jia: 
Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsE(E165Q)X/RipC complex in peptidisc
Method: single particle / : Li J, Xu X, Luo M

EMDB-34712: 
Cryo-EM structure of nucleotide-bound ComA at outward-facing state with EC gate closed conformation
Method: single particle / : Yu L, Xin X, Min L

EMDB-34713: 
Cryo-EM structure of nucleotide-bound ComA at outward-facing state with EC gate open conformation
Method: single particle / : Yu L, Xin X, Min L

EMDB-34714: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant
Method: single particle / : Yu L, Xin X, Min L

EMDB-34716: 
Cryo-EM structure of ComC bound ComA C17A at inward-facing state
Method: single particle / : Lin Y, Xin X, Min L

PDB-8hf4: 
Cryo-EM structure of nucleotide-bound ComA at outward-facing state with EC gate closed conformation
Method: single particle / : Yu L, Xin X, Min L

PDB-8hf5: 
Cryo-EM structure of nucleotide-bound ComA at outward-facing state with EC gate open conformation
Method: single particle / : Yu L, Xin X, Min L

PDB-8hf6: 
Cryo-EM structure of nucleotide-bound ComA E647Q mutant
Method: single particle / : Yu L, Xin X, Min L

EMDB-28036: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

PDB-8edm: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

EMDB-28035: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

EMDB-28037: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

EMDB-28038: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

PDB-8edl: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

PDB-8edn: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

PDB-8edo: 
Cryo-EM structure of the full-length human NF1 dimer
Method: single particle / : Darling JE, Merk A, Grisshammer R, Ognjenovic J

EMDB-17767: 
Cryo electron tomography of human choriocarcinoma cells
Method: electron tomography / : Tun WM, Yee NB-Y, Ho EML, Darrow MC, Basham M

EMDB-33864: 
Cryo-EM structure of C6-ceramide-bound SPT-ORMDL3 complex
Method: single particle / : Xie T, Liu P, Gong X

EMDB-33866: 
Cryo-EM structure of SPT-ORMDL3 complex
Method: single particle / : Xie T, Liu P, Gong X

EMDB-33868: 
Cryo-EM structure of SPT-ORMDL3 (ORMDL3-deltaN2) complex
Method: single particle / : Xie T, Liu P, Gong X

EMDB-33869: 
Cryo-EM structure of SPT-ORMDL3 (ORMDL3-N13A) complex
Method: single particle / : Xie T, Liu P, Gong X

PDB-7yiu: 
Cryo-EM structure of the C6-ceramide-bound SPT-ORMDL3 complex
Method: single particle / : Xie T, Liu P, Gong X

PDB-7yj1: 
Cryo-EM structure of SPT-ORMDL3 (ORMDL3-deltaN2) complex
Method: single particle / : Xie T, Liu P, Gong X

PDB-7yj2: 
Cryo-EM structure of SPT-ORMDL3 (ORMDL3-N13A) complex
Method: single particle / : Xie T, Liu P, Gong X

EMDB-35213: 
Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC/AmiB complex with ATP in peptidisc
Method: single particle / : Xu X, Li J, Luo M

EMDB-35201: 
Cryo-EM structure of Pseudomonas aeruginosa FtsE(WT)X/EnvC complex in peptidisc
Method: single particle / : Xu X, Li J, Luo M

EMDB-35203: 
Cryo-EM structure of Pseudomonas aeruginosa FtsE(WT)X complex in peptidisc
Method: single particle / : Xin X, Jianwei L, Min L

EMDB-35204: 
Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc
Method: single particle / : Xu X, Li J, Luo M

EMDB-35205: 
Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc
Method: single particle / : Xu X, Li J, Luo M

PDB-8i6o: 
Cryo-EM structure of Pseudomonas aeruginosa FtsE(WT)X/EnvC complex in peptidisc
Method: single particle / : Xu X, Li J, Luo M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
