-Search query
-Search result
Showing 1 - 50 of 57 items for (author: screaton & gr)
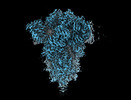
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
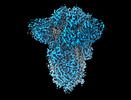
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-16680: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-15971: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-32839: 
CryoEM structure of sNS1 complexed with Fab5E3
Method: single particle / : Shu B, Lok SM

EMDB-32840: 
CryoEM structure of a dimer of loose sNS1 tetramer
Method: single particle / : Shu B, Lok SM

EMDB-32841: 
CryoEM structure of stable sNS1 tetramer
Method: single particle / : Shu B, Ooi JSG, Lok SM

EMDB-32842: 
CryoEM structure of loose sNS1 tetramer
Method: single particle / : Shu B, Lok SM

EMDB-32843: 
CryoEM structure of sNS1 hexamer
Method: single particle / : Shu B, Ooi JSG, Lok SM

EMDB-14885: 
OMI-42 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14886: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE RBD (local refinement)
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14887: 
OMI-2 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-14910: 
OMI-38 FAB IN COMPLEX WITH SARS-COV-2 BETA SPIKE
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-31677: 
DENV2_NGC_Fab_C10 28degree (1Fab:3E)
Method: single particle / : Shu B, Zhang S, Victor AK, Ng TS, Lok SM

EMDB-31678: 
DENV2_NGC_Fab_C10 28degrees (2Fab:3E)
Method: single particle / : Shu B, Zhang S, Victor AK, Ng TS, Lok SM

EMDB-31679: 
DENV2_NGC_Fab_C10 28degrees (3Fab:3E)
Method: single particle / : Shu B, Zhang S, Victor AK, Ng TS, Lok SM

EMDB-31680: 
DENV2_NGC_Fab_C10 4degrees (3Fab:3E)
Method: single particle / : Shu B, Zhang S, Victor AK, Ng TS, Lok SM

EMDB-13857: 
Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13868: 
Beta-50 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13869: 
COVOX-222 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13870: 
Beta-43 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13871: 
Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13872: 
Beta-32 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13873: 
Beta-53 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13874: 
Beta-44 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-13875: 
Beta-06 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-30465: 
Cryo-EM structure of dengue virus serotype 2 in complex with the scFv fragment of the broadly neutralizing antibody EDE1 C10
Method: single particle / : Zhang X, Sharma A, Duquerroy S, Zhou ZH, Rey FA

EMDB-12530: 
Subtomogram average of ChAdOx1 nCoV-19/AZD1222 derived SARS-CoV-2 spike glycoprotein
Method: subtomogram averaging / : Watanabe Y, Mendonca LM, Allen ER, Howe A, Lee M, Allen JD, Chawla H, Pulido D, Donnellan F, Davies H, Ulaszewska M, Belij-Rammerstorfer S, Morris S, Krebs AS, Dejnirattisai W, Mongkolsapaya J, Supasa P, Screaton GR, Green CM, Lambe T, Zhang P, Gilbert SC, Crispin M

EMDB-12274: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12275: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12276: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12277: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12278: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12279: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12280: 
EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-253H55L Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12281: 
EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-253H55L Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12282: 
EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-253H165L Fab
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12283: 
EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-12284: 
EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-159
Method: single particle / : Duyvesteyn HME, Zhao Y, Ren J, Stuart D

EMDB-11364: 
Cryo-EM structure of Spondweni virus prME
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11366: 
Cryo-EM structure of trimeric prME spike of Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11370: 
Cryo-EM structure of mature Dengue virus 2 at 3.1 angstrom resolution
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11371: 
Cryo-EM structure of mature Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

EMDB-11372: 
Cryo-EM structure of immature Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

PDB-6zqi: 
Cryo-EM structure of Spondweni virus prME
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

PDB-6zqj: 
Cryo-EM structure of trimeric prME spike of Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

PDB-6zqu: 
Cryo-EM structure of mature Dengue virus 2 at 3.1 angstrom resolution
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM

PDB-6zqv: 
Cryo-EM structure of mature Spondweni virus
Method: single particle / : Renner M, Dejnirattisai W, Carrique L, Serna Martin I, Karia D, Ilca SL, Ho SF, Kotecha A, Keown JR, Mongkolsapaya J, Screaton GR, Grimes JM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
