-Search query
-Search result
Showing 1 - 50 of 101 items for (author: scott & m & stagg)

EMDB-27593: 
The structure of S. epidermidis Cas10-Csm bound to target RNA
Method: single particle / : Paraan M, Stagg SM, Dunkle JA
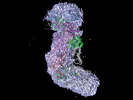
EMDB-27762: 
318 kDa Cas10-Csm effector complex bound to cognate target RNA
Method: single particle / : Paraan M, Stagg SM, Dunkle JA

PDB-8do6: 
The structure of S. epidermidis Cas10-Csm bound to target RNA
Method: single particle / : Paraan M, Stagg SM, Dunkle JA

EMDB-28147: 
Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

EMDB-28259: 
Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

EMDB-28269: 
Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

PDB-8ehg: 
Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

PDB-8emq: 
Mouse apoferritin heavy chain with zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

PDB-8en7: 
Mouse apoferritin heavy chain without zinc determined using single-particle cryo-EM with Apollo camera.
Method: single particle / : Peng R, Fu X, Mendez JH, Randolph PH, Bammes B, Stagg SM

EMDB-26172: 
Global average of aligned subtomograms of AAV2 bound with PKD1-2
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26173: 
First class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26174: 
Second class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26175: 
Third class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26176: 
Fourth class of AAV2 bound with PKD1-2 revealed by classification of aligned subtomgrams
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26177: 
SPR reconstruction of AAV5 bound with PKD1-2
Method: single particle / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26182: 
Global average of aligned subtomograms of AAV5 bound with PKD1-2
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26186: 
First class of AAV5 bound with PKD1-2 revealed by classification of aligned subtomgrams
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26187: 
Second class of AAV5 bound with PKD1-2 revealed by classification of aligned subtomgrams
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-26189: 
Third class of AAV5 bound with PKD1-2 revealed by classification of aligned subtomograms
Method: subtomogram averaging / : Hu GQ, Silveria MA, Chapman MS, Stagg SM

EMDB-20121: 
Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA
Method: single particle / : Hoffer ED, Mehrani A, Keiler KC, Stagg SM, Dunham CM

PDB-6om6: 
Structure of trans-translation inhibitor bound to E. coli 70S ribosome with P site tRNA
Method: single particle / : Hoffer ED, Mehrani A, Keiler KC, Stagg SM, Dunham CM

EMDB-21608: 
Mini-coat geometry for a clathrin coated vesicle: clathrin-focused
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21609: 
Mini-coat geometry for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21610: 
Basket geometry for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21611: 
Asymmetric vertex of the clathrin minicoat cage, subparticle refinement of the clathrin layer
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21612: 
Football geometry for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21613: 
Tennis geometry for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21614: 
Barrel geometry for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21615: 
Pentagonal face of the mini-coat cage for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-21616: 
Hexagonal face of the mini-coat cage for a clathrin coated vesicle
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

PDB-6wcj: 
Asymmetric vertex of the clathrin minicoat cage
Method: single particle / : Paraan M, Mendez J, Sharum S, Kurtin D, He H, Stagg S

EMDB-20905: 
Yeast RTP Interacts with Extended Termini of Clinet Protein Nop58p
Method: single particle / : Yu G, Zhao Y, Rai J, Stagg SM, Li H

EMDB-20155: 
Apoferritin from equine spleen at 2.9 Angstrom
Method: single particle / : Mendez JH, Randolph P, Stagg SM

EMDB-20156: 
Apoferritin from equine spleen at 3.9 Angstrom
Method: single particle / : Mendez JH, Randolph P, Stagg SM

EMDB-20157: 
Apoferritin from equine spleen at 4.3 Angstrom
Method: single particle / : Mendez JH, Randolph P, Stagg SM

EMDB-20158: 
Ribosome 70S at 2.8 Angstrom
Method: single particle / : Mehrani A, Mendez JH, Stagg SM

EMDB-0553: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2
Method: single particle / : Meyer NL, Xie Q, Davulcu O, Yoshioka C, Chapman MS

PDB-6nz0: 
Cryo-EM structure of AAV-2 in complex with AAVR PKD domains 1 and 2
Method: single particle / : Meyer NL, Xie Q, Davulcu O, Yoshioka C, Chapman MS

EMDB-0621: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: subtomogram averaging / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L
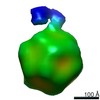
EMDB-0622: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: subtomogram averaging / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L

EMDB-0623: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: electron tomography / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L

EMDB-0624: 
Structure of the AAV2 with its Cell Receptor, AAVR
Method: electron tomography / : Hu GQ, Meyer NL, Stagg SM, Chapman MS, Davulcu O, Xie Q, Noble AJ, Yoshioka C, Gingerich D, Trzynka A, David L
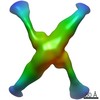
EMDB-7527: 
Class 1 vertex with 2-fold symmetry extracted from SEC13/SEC31deltaC cuboctahedrons and aligned, both using localized reconstruction method.
Method: single particle / : Paraan M, Bhattacharya N, Stagg S

EMDB-7333: 
Staphylococcus aureus phage 80alpha-derived SaPI1 mature capsid
Method: single particle / : Dokland T

PDB-6c21: 
Capsid protein in the Staphylococcus aureus phage 80alpha mature capsid
Method: single particle / : Kizziah JL, Dearborn AD, Dokland T

PDB-6c22: 
Capsid protein in the Staphylococcus aureus phage 80alpha-derived SaPI1 mature capsid
Method: single particle / : Kizziah JL, Dearborn AD, Dokland T

EMDB-8839: 
The Hsp90 Co-chaperon R2TP Forms a Hexameric Platform
Method: single particle / : Tian S, Yu G, He H, Liu P, Marshall A, Demeler B, Stagg S, Li H

EMDB-8574: 
Single particle reconstruction of chimeric adeno-associated virus-DJ with a Heparanoid Pentasaccharide
Method: single particle / : Xie Q, Noble AJ, Sousa DR, Meyer NL, Davulcu O, Zhang FM, Linhardt RJ, Stagg SM, Chapman MS

PDB-5uf6: 
The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide
Method: single particle / : Xie Q, Spear JM, Noble AJ, Sousa DR, Meyer NL, Davulcu O, Zhang F, Linhardt RJ, Stagg SM, Chapman M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
