-Search query
-Search result
Showing 1 - 50 of 88 items for (author: scott & dc)

EMDB-19856: 
Focused map 1- K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19857: 
Focused map 2 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19858: 
Focused map 3 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19859: 
Focused map 4 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-19860: 
Focused map 5 - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB~acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-18207: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide, Glacios map
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-18230: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA
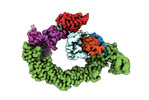
EMDB-18915: 
Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17798: 
CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17799: 
CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 2
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17800: 
NEDD8-CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 1
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17801: 
NEDD8-CUL2-RBX1-ELOB/C-FEM1C-SIL1 conformation 2
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17802: 
K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL1-RBX1-SKP1-FBXW7 with trapped UBE2R2-donor UB-acceptor UB-cyclin E peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17803: 
Consensus map - K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-17822: 
K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA

EMDB-18767: 
K48-linked ubiquitin chain formation with a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB~acceptor UB-BRD4
Method: single particle / : Liwocha J, Prabu JR, Kleiger G, Schulman BA
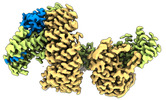
EMDB-14561: 
CAND1-CUL1-RBX1
Method: single particle / : Baek K, Schulman BA
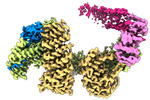
EMDB-14563: 
CAND1-SCF-SKP2 CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA
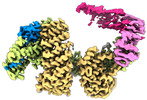
EMDB-14564: 
CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA
EMDB-14594: 
CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-14597: 
CAND1 delhairpin-SCF-SKP2 CAND1 partly engaged SCF partly rocked
Method: single particle / : Baek K, Schulman BA
EMDB-14598: 
CAND1 delhairpin-SCF-SKP2 CAND1 rolling SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14599: 
CAND1-SCF-FBXW7 CAND1 rolling-1a
Method: single particle / : Baek K, Schulman BA

EMDB-14600: 
CAND1-SCF-FBXW7 CAND1 rolling-1b
Method: single particle / : Baek K, Schulman BA

EMDB-14601: 
CAND1-SCF-FBXW7 CAND1 rolling-2c
Method: single particle / : Baek K, Schulman BA

EMDB-14602: 
CAND1-SCF-FBXW7 CAND1 rolling-2d
Method: single particle / : Baek K, Schulman BA
EMDB-14603: 
CAND1-SCF-FBXW7 CAND1 rolling-2a
Method: single particle / : Baek K, Schulman BA
EMDB-14604: 
CAND1-SCF-FBXW7 CAND1 rolling-2b
Method: single particle / : Baek K, Schulman BA

EMDB-14605: 
CAND1-SCF-FBXW7 CAND1 little CAND1
Method: single particle / : Baek K, Schulman BA
EMDB-14606: 
CAND1-SCF-FBXW7 CAND1 no CAND1-1
Method: single particle / : Baek K, Schulman BA

EMDB-14607: 
CAND1-SCF-FBXW7 CAND1 no CAND1-2
Method: single particle / : Baek K, Schulman BA
EMDB-14608: 
CAND1-SCF-FBXW7 (SKP1deldel) CAND1 engaged no SKP1-Fbp
Method: single particle / : Baek K, Schulman BA
EMDB-14609: 
CAND1-SCF-FBXW7 (SKP1deldel) CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA
EMDB-14610: 
CAND1delhairpin-SCF-FBXW7 CAND1 partly engaged SCF partly rocked
Method: single particle / : Baek K, Schulman BA

EMDB-14611: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2d SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-14612: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2c SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-14613: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2b SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-14614: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2a SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-14615: 
CAND1(2X)-SCF-FBXW7 no CAND1 SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14616: 
CAND1(2X)-SCF-FBXW7 CAND1 rolling-1 SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-16575: 
CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-16576: 
CAND1 b-hairpin++-SCF-SKP2 CAND1 partly engaged SCF partly rocked
Method: single particle / : Baek K, Schulman BA

EMDB-16577: 
CAND1(2X)-SCF-FBXW7 CAND1 rolling-2 SCF engaged
Method: single particle / : Baek K, Schulman BA
EMDB-16578: 
CAND1(2X)-SCF-FBXW7 CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA

EMDB-16579: 
CAND1-SCF-FBXW7 no CAND1 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA

EMDB-16580: 
CAND1-SCF-FBXW7 CAND1 rolling-1 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA
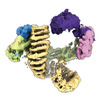
EMDB-16581: 
CAND1-SCF-FBXW7 CAND1 rolling-2 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA
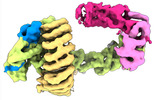
EMDB-16582: 
CAND1-SCF-SKP2 CAND1 rolling-1 SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-16583: 
CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA
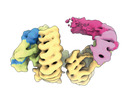
EMDB-16584: 
CAND1-SCF-SKP2 CAND1 engaged SCF rocked (low res)
Method: single particle / : Baek K, Schulman BA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
