-Search query
-Search result
Showing all 29 items for (author: scott & c & weaver)

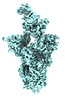
EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS
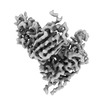
EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS
EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tm1: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tma: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS

PDB-6djy: 
Fako virus
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7941: 
Fako virus empty particles aligned to the best decoy map (asymmetric reconstruction) showing the locations of the polymerase complexes
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7944: 
Fako virus full particles, icosahedral reconstruction
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7945: 
Fako virus empty particles, icosahedral reconstruction
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7948: 
Fako virus empty particles, D2-symmetrized reconstruction
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7949: 
Fako virus full particles, D2-symmetrized reconstruction
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7953: 
Fako virus empty particles, projection-matching reconstruction without imposed symmetry
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7954: 
Fako virus full particles, projection-matching reconstruction without imposed symmetry
Method: single particle / : Kaelber JT, Jiang W, Weaver SC, Auguste AJ, Chiu W

EMDB-7513: 
Subtomogram average of mature CHIKV virion budded from human cells
Method: subtomogram averaging / : Galaz-Montoya JG, Jin J, Sherman MB

EMDB-7515: 
Subtomogram averages of budding-arrested nucleocapsid-like particles (NCLPs) inside CHIKV-infected human cells.
Method: subtomogram averaging / : Galaz-Montoya JG, Jin J, Sherman MB

EMDB-7003: 
Eilat virus/Venezuelan equine encephalitis virus chimeric vaccine candidate
Method: single particle / : Kaelber JT, Erasmus JH, Weaver SC, Nasar F, Chiu W

EMDB-7004: 
Eilat virus/eastern equine encephalitis virus chimeric vaccine candidate
Method: single particle / : Sherman MB, Erasmus JH, Frolov I, Weaver SC, Nasar F

EMDB-3406: 
Eilat virus/Chikungunya virus chimeric vaccine candidate
Method: single particle / : Kaelber JT, Erasmus JH, Kim DY, Frolov I, Nasar F, Weaver SC, Chiu W

EMDB-8071: 
Map of Venezuelan Equine Encephalitis Virus by cryoSPT (subtomogram averaging)
Method: subtomogram averaging / : Galaz-Montoya JG

EMDB-6001: 
Fako virus capsid: a newly-isolated reovirus has the simplest genomic and structural organization of any reovirus
Method: single particle / : Auguste AJ, Kaelber JT, Fokam EB, Guzman H, Carrington CVF, Erasmus JH, Kamgang B, Popov VL, Jakana J, Liu X, Wood TG, Widen SG, Vasilakis N, Tesh RB, Chiu W, Weaver SC

EMDB-6002: 
Fako virus virion: a newly-isolated reovirus has the simplest genomic and structural organization of any reovirus
Method: single particle / : Auguste AJ, Kaelber JT, Fokam EB, Guzman H, Carrington CVF, Erasmus JH, Kamgang B, Popov VL, Jakana J, Liu X, Wood TG, Widen SG, Vasilakis N, Tesh RB, Chiu W, Weaver SC

EMDB-5563: 
Electron microscopy of Everglades virus
Method: single particle / : Sherman MB, Trujillo J, Leahy I, Razmus D, DeHate R, Lorcheim P, Czarneski MA, Zimmerman D, Newton JPM, Haddow AM, Weaver SC

PDB-2yew: 
Modeling Barmah Forest virus structural proteins
Method: single particle / : Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, Lok SM

EMDB-1886: 
The 6A cryo-EM reconstruction of Barmah Forest virus
Method: single particle / : Kostyuchenko VA, Jakana J, Liu X, Haddow AD, Aung M, Weaver SC, Chiu W, Lok SM

PDB-3j0c: 
Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map
Method: single particle / : Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W

PDB-3j0g: 
Homology model of E3 protein of Venezuelan Equine Encephalitis Virus TC-83 strain fitted with a cryo-EM map
Method: single particle / : Zhang R, Hryc CF, Chiu W

EMDB-5275: 
4.4 Angstrom Cryo-EM Structure of an Enveloped Alphavirus Venezuelan Equine Encephalitis Virus
Method: single particle / : Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W

EMDB-5276: 
4.4 Angstrom Cryo-EM Structure of an Enveloped Alphavirus Venezuelan Equine Encephalitis Virus
Method: single particle / : Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W

EMDB-5210: 
The structure of the recombinant alphavirus, Western equine encephalitis virus, revealed by cryoelectron microscopy
Method: single particle / : Sherman MB, Weaver SC
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
