-Search query
-Search result
Showing 1 - 50 of 202 items for (author: schoehn & g)

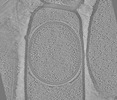
EMDB-19405: 
Tomograms of spoIVB Bacillus subtilis sporangia
Method: electron tomography / : Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C
EMDB-19411: 
Tomograms of cotE Bacillus subtilis sporangia
Method: electron tomography / : Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C

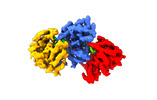
EMDB-18043: 
Helical structure of the influenza A virus ribonucleoprotein-like
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T
EMDB-18044: 
Focused reconstruction of influenza A RNP-like particle
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T

PDB-8pzp: 
Model for influenza A virus helical ribonucleoprotein-like structure
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T

PDB-8pzq: 
Model for focused reconstruction of influenza A RNP-like particle
Method: helical / : Chenavier F, Estrozi LF, Zarkadas E, Ruigrok RWH, Schoehn G, Ballandras-Colas A, Crepin T

EMDB-14863: 
Bacteriophage T5 head - pb10-N-Ter fused to OVA
Method: single particle / : Schoehn G, Boulanger P, Benihoud K

EMDB-15967: 
Cryo-EM structure of the proximal end of bacteriophage T5 tail : p142 tail terminator protein hexamer and pb6 tail tube protein trimer
Method: single particle / : Linares R, Effantin G, Breyton C, Darnault C, Epalle N, Boeri Erba E, Schoehn G

EMDB-15968: 
Cryo-EM structure of the proximal end of bacteriophage T5 tail, after interaction with its receptor : p142 tail terminator protein hexamer and pb6 tail tube protein trimer
Method: single particle / : Linares R, Effantin G, Breyton C, Darnault C, Epalle N, Boeri Erba E, Schoehn G

PDB-8cpc: 
3D electron diffraction structure of Hen Egg-White Lysozyme from nano-crystals obtained by high pressure freezing and cryo-sectioning
Method: electron crystallography / : Moriscot C, Schoehn G, Housset D

EMDB-16687: 
Carin1 bacteriophage mature capsid
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

EMDB-16688: 
Carin1 bacteriophage portal assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

EMDB-16689: 
Carin 1 bacteriophage tail, connector and tail fibers assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

PDB-8cjz: 
Carin1 bacteriophage mature capsid
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

PDB-8ck0: 
Carin1 bacteriophage portal assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

PDB-8ck1: 
Carin 1 bacteriophage tail, connector and tail fibers assembly
Method: single particle / : d'Acapito A, Neumann E, Schoehn G

EMDB-14733: 
Tail tip of siphophage T5 : tip proteins
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-14790: 
Tail tip of siphophage T5 : central fibre
Method: single particle / : Linares R, Arnaud CA, Effantin G, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-14799: 
Tail tip of siphophage T5 : full complex after interaction with its bacterial receptor FhuA
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-14800: 
Tail tip of siphophage T5 : bent fibre after interaction with its bacterial receptor FhuA
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-14869: 
Tail tip of siphophage T5 : full structure
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-14873: 
Tail tip of siphophage T5 : open cone after interaction with bacterial receptor FhuA
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7zhj: 
Tail tip of siphophage T5 : tip proteins
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7zlv: 
Tail tip of siphophage T5 : central fibre protein pb4
Method: single particle / : Linares R, Arnaud CA, Effantin G, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7zn2: 
Tail tip of siphophage T5 : full complex after interaction with its bacterial receptor FhuA
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7zn4: 
Tail tip of siphophage T5 : bent fibre after interaction with its bacterial receptor FhuA
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7zqb: 
Tail tip of siphophage T5 : full structure
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7zqp: 
Tail tip of siphophage T5 : open cone after interaction with bacterial receptor FhuA
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-15802: 
T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA
Method: single particle / : Degroux S, Effantin G, Linares R, Schoehn G, Breyton C

PDB-8b14: 
T5 Receptor Binding Protein pb5 in complex with its E. coli receptor FhuA
Method: single particle / : Degroux S, Effantin G, Linares R, Schoehn G, Breyton C

EMDB-15145: 
Leafy, UFO, ASK1 and DNA complex
Method: single particle / : Rieu P, Turchi L, Thevenon E, Zarkadas E, Nanao M, Chahtane H, Tichtinsky G, Lucas J, Blanc-Mathieu R, Zubieta C, Schoehn G, Parcy F

EMDB-16435: 
microtubule decorated with tubulin oligomers in presence of APC C-terminal domain. (here only the microtubule map section is represented)
Method: helical / : Serre L, Arnal I

PDB-8c5c: 
microtubule decorated with tubulin oligomers in presence of APC C-terminal domain. (here only map corresponding to the 13-pf microtubule is represented)
Method: helical / : Serre L, Arnal I

EMDB-16436: 
microtubule decorated with tubulin oligomers in presence of APC C-terminal domain. (here only map corresponding to the external tubulin oligomer is represented)
Method: helical / : Serre L, Arnal I

EMDB-14630: 
Membrane-bound CHMP2A-CHMP3 filament (430 Angstrom diameter)
Method: helical / : Azad K, Desfosses A, Effantin G, Schoehn G, Weissenhorn W

EMDB-14631: 
Membrane-bound CHMP2A-CHMP3 filament (410 Angstrom diameter)
Method: helical / : Azad K, Desfosses A, Effantin G, Schoehn G, Weissenhorn W

PDB-7zcg: 
CHMP2A-CHMP3 heterodimer (430 Angstrom diameter)
Method: helical / : Azad K, Desfosses A, Effantin G, Schoehn G, Weissenhorn W

PDB-7zch: 
CHMP2A-CHMP3 heterodimer (410 Angstrom diameter)
Method: helical / : Azad K, Desfosses A, Effantin G, Schoehn G, Weissenhorn W

EMDB-13953: 
Tail tip of siphophage T5 : common core proteins
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

PDB-7qg9: 
Tail tip of siphophage T5 : common core proteins
Method: single particle / : Linares R, Arnaud CA, Effantin G, Darnault C, Epalle N, Boeri Erba E, Schoehn G, Breyton C

EMDB-14582: 
Human heparan sulfate polymerase complex EXT1-EXT2
Method: single particle / : Leisico F, Omeiri J, Hons M, Schoehn G, Lortat-Jacob H, Wild R

PDB-7zay: 
Human heparan sulfate polymerase complex EXT1-EXT2
Method: single particle / : Leisico F, Omeiri J, Hons M, Schoehn G, Lortat-Jacob H, Wild R

EMDB-4928: 
Cryo-EM reconstruction of TMV coat protein
Method: helical / : Kandiah E, Effantin G

EMDB-15574: 
Symmetric hexamer of vaccinia virus DNA helicase D5 residues 323-785
Method: single particle / : Burmeister WP, Hutin S, Ling WL, Grimm C, Schoehn G

EMDB-15575: 
Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C1
Method: single particle / : Burmeister WP, Hutin S, Ling WL, Grimm C, Schoehn G

PDB-8apl: 
Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C6
Method: single particle / : Burmeister WP, Hutin S, Ling WL, Grimm C, Schoehn G

PDB-8apm: 
Vaccinia virus DNA helicase D5 residues 323-785 hexamer with bound DNA processed in C1
Method: single particle / : Burmeister WP, Hutin S, Ling WL, Grimm C, Schoehn G

EMDB-12775: 
Cryo-EM structure of the plectasin fibril (double strands)
Method: helical / : Effantin G

EMDB-12776: 
Cryo-EM structure of the plectasin fibril (single strand)
Method: helical / : Effantin G

PDB-7oae: 
Cryo-EM structure of the plectasin fibril (double strands)
Method: helical / : Effantin G
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
