-Search query
-Search result
Showing 1 - 50 of 767 items for (author: schmid & m)
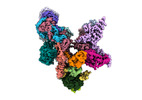
EMDB-18496: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18499: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map A
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18500: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map B.
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18502: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map D
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18503: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba.- Map E
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18504: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map F
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

PDB-8qma: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS

EMDB-18892: 
Lipid droplet-Vacuole contacts in Ldo16 overexpression yeast strain.
Method: electron tomography / : Collado J

EMDB-18893: 
Lipid droplet-vacuole and Nucleus-vacuole contacts in WT yeast cell starved for 4 hours
Method: electron tomography / : Collado J

EMDB-18894: 
Lipid droplet lipophagy in 4-hour starved WT yeast cell.
Method: electron tomography / : Collado J

EMDB-18895: 
Multiple vacuole-lipid droplet-nucleus contacts in 4-hour starved WT yeast cell.
Method: electron tomography / : Collado J

EMDB-18896: 
Lipophagy in 5-day starved WT yeast cell.
Method: electron tomography / : Collado J

EMDB-18897: 
Lipid droplets in proximity to a vacuole in dLdo strain cell after 5-day starvation.
Method: electron tomography / : Collado J

EMDB-18898: 
Vacuolar contents of WT cell after 5-day starvation.
Method: electron tomography / : Collado J

EMDB-18899: 
Lipid droplet-nucleus contacts in dLdo yeast strain after 5-day starvation.
Method: electron tomography / : Collado J
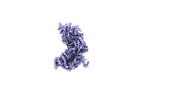
EMDB-18501: 
Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba - Map C
Method: single particle / : do Prado PFV, Ahrens FM, Pfannschmidt T, Hillen HS
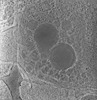
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
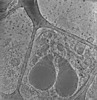
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-17104: 
AApoAII amyloid fibril Morphology II (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

EMDB-17105: 
AApoAII amyloid fibril Morphology I (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

PDB-8oq4: 
AApoAII amyloid fibril Morphology II (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

PDB-8oq5: 
AApoAII amyloid fibril Morphology I (ex vivo)
Method: helical / : Andreotti G, Schmidt M, Faendrich M

EMDB-29738: 
Influenza A H3N2 X-31 Hemagglutinin in complex with FL-1061
Method: single particle / : Windsor IW, Thornlow D, Schmidt AG

PDB-8g5b: 
Influenza A H3N2 X-31 Hemagglutinin in complex with FL-1061
Method: single particle / : Windsor IW, Thornlow D, Schmidt AG

EMDB-29737: 
X-31 hemagglutinin in complex with FL-1061 Fab
Method: single particle / : Windsor IW, Thornlow D, Schmidt AG

PDB-8g5a: 
X-31 hemagglutinin in complex with FL-1061 Fab
Method: single particle / : Windsor IW, Thornlow D, Schmidt AG

EMDB-17166: 
unseeded Abeta(1-40) amyloid fibril (morphology i)
Method: helical / : Pfeiffer PB, Schmidt M, Faendrich M

EMDB-17167: 
unseeded Abeta(1-40) amyloid fibril (morphology ii)
Method: helical / : Pfeiffer PB, Schmidt M, Faendrich M

EMDB-17168: 
seeded Abeta(1-40) amyloid fibril (morphology I)
Method: helical / : Pfeiffer PB, Schmidt M, Faendrich M

PDB-8ot1: 
unseeded Abeta(1-40) amyloid fibril (morphology i)
Method: helical / : Pfeiffer PB, Schmidt M, Faendrich M

PDB-8ot3: 
unseeded Abeta(1-40) amyloid fibril (morphology ii)
Method: helical / : Pfeiffer PB, Schmidt M, Faendrich M

PDB-8ot4: 
seeded Abeta(1-40) amyloid fibril (morphology I)
Method: helical / : Pfeiffer PB, Schmidt M, Faendrich M

EMDB-18715: 
TDP-43 amyloid fibrils: Morphology-1a
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

EMDB-18716: 
TDP-43 amyloid fibrils: Morphology-1b
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

EMDB-18717: 
TDP-43 amyloid fibrils: Morphology-2
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

PDB-8qx9: 
TDP-43 amyloid fibrils: Morphology-1a
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

PDB-8qxa: 
TDP-43 amyloid fibrils: Morphology-1b
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

PDB-8qxb: 
TDP-43 amyloid fibrils: Morphology-2
Method: helical / : Sharma K, Shenoy J, Loquet A, Schmidt M, Faendrich M

EMDB-29395: 
Subtomogram average of HuCoV-NL63 spike protein from purified intact virions
Method: subtomogram averaging / : Chen M, Chmielewski D, Schmid M, Jin J, Chiu W

EMDB-15214: 
Endogenous yeast L-A helper virus identified from native cell extracts
Method: single particle / : Schmidt L, Kyrilis F, Hamdi F, Semchonok DA, Kastritis PL

EMDB-15189: 
Capsid structure of the L-A helper virus from native viral communities
Method: single particle / : Schmidt L, Tueting C, Kyrilis F, Hamdi F, Semchonok DA, Kastritis PL

EMDB-15215: 
Asymmetric reconstruction of averaged ribosomes from Saccharomyces cerevisiae
Method: single particle / : Schmidt L, Tueting C, Kyrilis F, Hamdi F, Semchonok DA, Kastritis PL

PDB-8a5t: 
Capsid structure of the L-A helper virus from native viral communities
Method: single particle / : Schmidt L, Tueting C, Stubbs MT, Kastritis PL

EMDB-17736: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17737: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17738: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pke: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkf: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkg: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
