-Search query
-Search result
Showing 1 - 50 of 65 items for (author: scharf & l)
EMDB-29383: 
Structure of baseplate with receptor binding complex of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fqc: 
Structure of baseplate with receptor binding complex of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH
EMDB-29353: 
Structure of Agrobacterium tumefaciens bacteriophage Milano curved tail
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-29354: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-tube
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-29355: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-sheath
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fop: 
Structure of Agrobacterium tumefaciens bacteriophage Milano curved tail
Method: single particle / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8fou: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-tube
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

PDB-8foy: 
Structure of Agrobacterium tumefaciens bacteriophage Milano contracted tail-sheath
Method: helical / : Sonani RR, Leiman PG, Wang F, Kreutzberger MAB, Sebastian A, Esteves NC, Kelly RJ, Scharf B, Egelman EH

EMDB-29500: 
Portal assembly of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH
EMDB-29501: 
Collar sheath structure of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-29503: 
Neck structure of Agrobacterium phage Milano, C3 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH
EMDB-29504: 
Structure of neck and portal vertex of Agrobacterium phage Milano, C5 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-29512: 
Structure of tail-neck junction of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-29540: 
Structure of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH
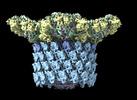
EMDB-29541: 
Structure of neck with portal vertex of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwb: 
Portal assembly of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwc: 
Collar sheath structure of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwe: 
Neck structure of Agrobacterium phage Milano, C3 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwg: 
Structure of neck and portal vertex of Agrobacterium phage Milano, C5 symmetry
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fwm: 
Structure of tail-neck junction of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fxp: 
Structure of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

PDB-8fxr: 
Structure of neck with portal vertex of capsid of Agrobacterium phage Milano
Method: single particle / : Sonani RR, Wang F, Esteves NC, Kelly RJ, Sebastian A, Kreutzberger MAB, Leiman PG, Scharf BE, Egelman EH

EMDB-26995: 
Cryo-EM helical reconstruction of the EPEC H6 Curly I flagellar core
Method: helical / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

EMDB-27008: 
Cryo-EM structure of the supercoiled EPEC H6 flagellar filament core Curly I waveform
Method: single particle / : Kreutzberger MA, Chatterjee S, Frankel G, Egelman EH

EMDB-27026: 
Cryo-EM structure of the supercoiled S. islandicus REY15A archaeal flagellar filament
Method: single particle / : Kreutzberger MAB, Liu J, Krupovic M, Egelman EH

EMDB-27029: 
Longer Cryo-EM Density map of the EPEC H6 Curly I bacterial flagellar filament
Method: single particle / : Kreutzberger MA, Chatterjee S, Frankel G, Egelman EH

EMDB-27059: 
Cryo-EM structure of the helical E. coli K12 flagellar filament core
Method: helical / : Sonani RR, Kreutzberger MAB, Sebastian AL, Scharf B, Egelman EH

EMDB-27060: 
Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform
Method: single particle / : Sonani RR, Kreutzberger MAB, Sebastian AL, Scharf B, Egelman EH

EMDB-27064: 
Cryo-EM Helical Reconstruction of the EPEC H6 flagellar filament in the Normal waveform
Method: helical / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

EMDB-27065: 
Cryo-EM helical reconstruction of the S. islandicus REY15A archaeal flagellar filament
Method: helical / : Kreutzberger MAB, Liu J, Krupovic M, Egelman EH

EMDB-27076: 
Cryo-EM asymmetric reconstruction of the EPEC H6 bacterial flagellar filament Normal Waveform
Method: single particle / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

PDB-8cvi: 
Cryo-EM structure of the supercoiled EPEC H6 flagellar filament core Curly I waveform
Method: single particle / : Kreutzberger MA, Chatterjee S, Frankel G, Egelman EH

PDB-8cwm: 
Cryo-EM structure of the supercoiled S. islandicus REY15A archaeal flagellar filament
Method: single particle / : Kreutzberger MAB, Liu J, Krupovic M, Egelman EH

PDB-8cxm: 
Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform
Method: single particle / : Sonani RR, Kreutzberger MAB, Sebastian AL, Scharf B, Egelman EH

PDB-8cye: 
Cryo-EM asymmetric reconstruction of the EPEC H6 bacterial flagellar filament Normal Waveform
Method: single particle / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

EMDB-25211: 
Cryo-EM structure of the enterohemorrhagic E. coli O157:H7 flagellar filament
Method: helical / : Kreutzberger MAB, Wang F, Egelman EH

EMDB-25212: 
Cryo-EM structure of the N319F, N323F EHEC O157:H7 flagellar filament
Method: helical / : Kreutzberger MAB, Sauder AB, Kendall MM, Egelman EH

EMDB-25213: 
Cryo-EM structure of the enteropathogenic E. coli O127:H6 flagellar filament
Method: single particle / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

EMDB-25215: 
Cryo-EM structure of the Sinorhizobium meliloti flagellar filament
Method: helical / : Kreutzberger MAB, Scharf BE, Egelman EH

EMDB-25382: 
Cryo-EM structure of the Achromobacter flagellar filament
Method: helical / : Kreutzberger MA, Wang F, Egelman EH

EMDB-25386: 
Cryo-EM structure of the seam subunits of the enteropathogenic E. coli O127:H6 flagellar filament
Method: single particle / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

EMDB-25388: 
Cryo-EM map of the EHEC O157:H7 flagellar filament outer domain sheath with D1 symmetry
Method: helical / : Kreutzberger MAB, Wang F, Egelman EH

PDB-7sn4: 
Cryo-EM structure of the enterohemorrhagic E. coli O157:H7 flagellar filament
Method: helical / : Kreutzberger MAB, Wang F, Egelman EH

PDB-7sn7: 
Cryo-EM structure of the enteropathogenic E. coli O127:H6 flagellar filament
Method: single particle / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

PDB-7sn9: 
Cryo-EM structure of the Sinorhizobium meliloti flagellar filament
Method: helical / : Kreutzberger MAB, Scharf BE, Egelman EH

PDB-7sqd: 
Cryo-EM structure of the Achromobacter flagellar filament
Method: helical / : Kreutzberger MA, Wang F, Egelman EH

PDB-7sqj: 
Cryo-EM structure of the seam subunits of the enteropathogenic E. coli O127:H6 flagellar filament
Method: single particle / : Kreutzberger MAB, Chatterjee S, Frankel G, Egelman EH

EMDB-31456: 
Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex
Method: single particle / : Zhang H, Chen L, Mao C, Shen Q, Yang D, Shen D, Qin J

EMDB-31457: 
Cryo-EM structure of afamelanotide-MC4R-Gs_Nb35 complex
Method: single particle / : Zhang H, Chen L, Mao C, Shen Q, Yang D, Shen D, Qin J

EMDB-31458: 
Cryo-EM structure of bremelanotide-MC4R-Gs_Nb35 complex
Method: single particle / : Zhang H, Chen L, Mao C, Shen Q, Yang D, Shen D, Qin J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
