-Search query
-Search result
Showing all 32 items for (author: sashital & dg)

EMDB-27159: 
Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/PAM prespacer
Method: single particle / : Dhingra Y, Suresh SK, Juneja P, Sashital DG

EMDB-27160: 
Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/Processed prespacer
Method: single particle / : Dhingra Y, Suresh SK, Juneja P, Sashital DG

EMDB-27161: 
Type I-C Cas4-Cas1-Cas2 complex bound to half-site integration intermediate (HSI)
Method: single particle / : Dhingra Y, Suresh SK, Juneja P, Sashital DG

EMDB-27162: 
Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/NoPAM prespacer
Method: single particle / : Dhingra Y, Suresh SK, Juneja P, Sashital DG

EMDB-27769: 
Type I-C Cas4-Cas1-Cas2 complex bound to a 24 bp duplex PAM/PAM prespacer
Method: single particle / : Dhingra Y, Suresh SK, Juneja P, Sashital DG

EMDB-20127: 
Bacillus halodurans Cas1-Cas2 complex in presence of target CRISPR DNA
Method: single particle / : Sashital DG, Lee H, Dhingra Y

EMDB-20128: 
Bacillus halodurans Cas4-Cas1-Cas2 asymmetrical complex in presence of target CRISPR DNA
Method: single particle / : Sashital DG, Lee H, Dhingra Y

EMDB-20129: 
Bacillus halodurans Cas4-Cas1-Cas2 symmetrical complex in presence of target CRISPR DNA
Method: single particle / : Sashital DG, Lee H, Dhingra Y

EMDB-20130: 
Bacillus halodurans Cas4-Cas1-Cas2 asymmetrical complex in presence of prespacer DNA
Method: single particle / : Sashital DG, Lee H, Dhingra Y

EMDB-20131: 
Bacillus halodurans Cas4-Cas1-Cas2 symmetrical complex in presence of prespacer DNA
Method: single particle / : Sashital DG, Lee H, Dhingra Y

EMDB-7485: 
Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays
Method: single particle / : Lee H, Zhou Y, Taylor DW, Sashital DG

EMDB-6125: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6126: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6127: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6128: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6129: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6130: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6131: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6132: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6133: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR
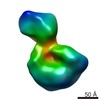
EMDB-6134: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6135: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6136: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6137: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6138: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6139: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6140: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6141: 
Negative stain random conical tilt reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6142: 
Cryo-EM reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6143: 
Cryo-EM reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR
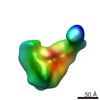
EMDB-6144: 
Cryo-EM reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR

EMDB-6145: 
Cryo-EM reconstructions of E. coli ribosomal 30S subunit assembly intermediates
Method: single particle / : Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
