-Search query
-Search result
Showing all 42 items for (author: rodrigues & p)

EMDB-19405: 
Tomograms of spoIVB Bacillus subtilis sporangia
Method: electron tomography / : Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C
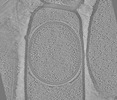
EMDB-19411: 
Tomograms of cotE Bacillus subtilis sporangia
Method: electron tomography / : Bauda E, Gallet B, Moravcova J, Effantin G, Chan H, Novacek J, Jouneau PH, Rodrigues CDA, Schoehn G, Moriscot C, Morlot C

EMDB-42480: 
Cryo-EM reconstruction of Staphylococcus aureus Oleate hydratase (OhyA) dimer with an ordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

EMDB-42484: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer with a disordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

PDB-8ur3: 
Cryo-EM reconstruction of Staphylococcus aureus Oleate hydratase (OhyA) dimer with an ordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

PDB-8ur6: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer with a disordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ
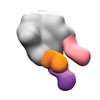
EMDB-40242: 
BG505 MD39 SOSIP in complex with Rh.NJ85 wk12 gp120GH, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40243: 
BG505 MD39 SOSIP in complex with Rh.NJ86 wk12 V1V3, C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40244: 
BG505 MD39 SOSIP in complex with Rh.NJ76 wk12 C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB
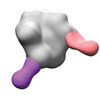
EMDB-40252: 
BG505 MD39 SOSIP in complex with Rh.NK04 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40254: 
BG505 MD39 SOSIP in complex with Rh.NJ75 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40255: 
BG505 MD39 SOSIP in complex with Rh.NJ87 wk12 C3V5 and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40256: 
BG505 MD39 SOSIP in complex with Rh.NJ84 wk12 V1V3, gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40257: 
BG505 MD39 SOSIP in complex with Rh.NJ77 wk12 V1V3, C3V5, N611/FP and base epitope polyclonal
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB
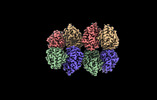
EMDB-28013: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)
Method: single particle / : Ambrosio AL, Dias SM, Quesnay JE, Portugal RV, Cassago A, van Heel MG, Islam Z, Rodrigues CT

PDB-8ec6: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)
Method: single particle / : Ambrosio AL, Dias SM, Quesnay JE, Portugal RV, Cassago A, van Heel MG, Islam Z, Rodrigues CT

EMDB-16311: 
Structure of the rod CNG channel bound to calmodulin
Method: single particle / : Marino J

EMDB-16313: 
The structure of the rod CNG channel bound to calmodulin
Method: single particle / : Marino J

EMDB-15987: 
In situ sub-tomogram average of the Ca. L. ossiferum ribosome
Method: subtomogram averaging / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15988: 
In situ sub-tomogram average of Ca. L. ossiferum actin filament
Method: subtomogram averaging / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15989: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15990: 
cryo-tomogram of Ca. L. ossiferum protrusions
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15991: 
cryo-tomogram of a Ca.L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15992: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15993: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-21705: 
PL-bound rat TRPV2 in nanodiscs
Method: single particle / : Pumroy RP, Moiseenkova-Bell VY

PDB-6wkn: 
PL-bound rat TRPV2 in nanodiscs
Method: single particle / : Pumroy RP, Moiseenkova-Bell VY

EMDB-11275: 
MreC
Method: helical / : Estrozi LF, Contreras-Martel C

PDB-6zlv: 
MreC
Method: helical / : Estrozi LF, Contreras-Martel C

EMDB-21983: 
Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis
Method: single particle / : Tan YZ, Rodrigues J

PDB-6x0o: 
Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis
Method: single particle / : Tan YZ, Rodrigues J, Keener JE, Zheng RB, Brunton R, Kloss B, Giacometti SI, Rosario AL, Zhang L, Niederweis M, Clarke OB, Lowary TL, Marty MT, Archer M, Potter CS, Carragher B, Mancia F

EMDB-21580: 
Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria
Method: single particle / : Tan YZ, Zhang L, Rodrigues J, Zheng RB, Giacometti SI, Rosario AL, Kloss B, Dandey VP, Wei H, Brunton R, Raczkowski AM, Athayde D, Catalao MJ, Pimentel M, Clarke OB, Lowary TL, Archer M, Niederweis M, Potter CS, Carragher B, Mancia F

EMDB-21600: 
Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1
Method: single particle / : Tan YZ, Zhang L, Rodrigues J, Zheng RB, Giacometti SI, Rosario AL, Kloss B, Dandey VP, Wei H, Brunton R, Raczkowski AM, Athayde D, Catalao MJ, Pimentel M, Clarke OB, Lowary TL, Archer M, Niederweis M, Potter CS, Carragher B, Mancia F

EMDB-21601: 
Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2
Method: single particle / : Tan YZ, Zhang L, Rodrigues J, Zheng RB, Giacometti SI, Rosario AL, Kloss B, Dandey VP, Wei H, Brunton R, Raczkowski AM, Athayde D, Catalao MJ, Pimentel M, Clarke OB, Lowary TL, Archer M, Niederweis M, Potter CS, Carragher B, Mancia F

PDB-6w98: 
Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria
Method: single particle / : Tan YZ, Zhang L, Rodrigues J, Zheng RB, Giacometti SI, Rosario AL, Kloss B, Dandey VP, Wei H, Brunton R, Raczkowski AM, Athayde D, Catalao MJ, Pimentel M, Clarke OB, Lowary TL, Archer M, Niederweis M, Potter CS, Carragher B, Mancia F

PDB-6wbx: 
Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1
Method: single particle / : Tan YZ, Zhang L, Rodrigues J, Zheng RB, Giacometti SI, Rosario AL, Kloss B, Dandey VP, Wei H, Brunton R, Raczkowski AM, Athayde D, Catalao MJ, Pimentel M, Clarke OB, Lowary TL, Archer M, Niederweis M, Potter CS, Carragher B, Mancia F

PDB-6wby: 
Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2
Method: single particle / : Tan YZ, Zhang L, Rodrigues J, Zheng RB, Giacometti SI, Rosario AL, Kloss B, Dandey VP, Wei H, Brunton R, Raczkowski AM, Athayde D, Catalao MJ, Pimentel M, Clarke OB, Lowary TL, Archer M, Niederweis M, Potter CS, Carragher B, Mancia F
EMDB-4072: 
Cryo-EM 3D reconstruction of rings formed by the extracellular domain of SpoIIIAG from Bacillus subtilis
Method: single particle / : Rodrigues C, Henry X, Neumann E, Schoehn G, Rudner D, Morlot C

EMDB-3222: 
Structure of a Chaperone-Usher pilus reveals the molecular basis of rod uncoilin
Method: helical / : Hospenthal MK, Redzej A, Dodson K, Ukleja M, Frenz B, Hultgren SJ, DiMaio F, Egelman EH, Waksman G

PDB-5flu: 
Structure of a Chaperone-Usher pilus reveals the molecular basis of rod uncoilin
Method: helical / : Hospenthal MK, Redzej A, Dodson K, Ukleja M, Frenz B, Hultgren SJ, DiMaio F, Egelman EH, Waksman G

EMDB-2929: 
Mechanism of Ubiquitin Ligation to a Substrate by human Anaphase-Promoting Complex
Method: single particle / : Brown N, VanderLinden R, Watson E, Qiao R, Grace CR, Yamaguchi M, Weissmann F, Frye J, Dube P, Cho SE, Actis M, Rodrigues P, Fujii N, Peters JM, Schulman BA, Stark H
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
