-Search query
-Search result
Showing 1 - 50 of 76 items for (author: roberts & ga)

EMDB-19132: 
Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Mladenov M, Seda M, Jenkins D, Stephens DJ, Roberts AJ

EMDB-19133: 
Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Seda M, Jenkins D, Stephens DJ, Roberts AJ

PDB-8rgg: 
Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Mladenov M, Seda M, Jenkins D, Stephens DJ, Roberts AJ

PDB-8rgh: 
Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1.
Method: single particle / : Mukhopadhyay AG, Toropova K, Daly L, Wells J, Vuolo L, Seda M, Jenkins D, Stephens DJ, Roberts AJ

EMDB-40914: 
Cryo-EM structure of cinacalcet-bound active-state human calcium-sensing receptor CaSR in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G
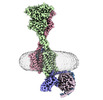
EMDB-40915: 
Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

EMDB-40916: 
Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G
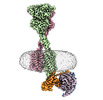
EMDB-40917: 
Cryo-EM structure of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

PDB-8szf: 
Cryo-EM structure of cinacalcet-bound active-state human calcium-sensing receptor CaSR in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

PDB-8szg: 
Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

PDB-8szh: 
Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G

PDB-8szi: 
Cryo-EM structure of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs
Method: single particle / : He F, Wu C, Gao Y, Skiniotis G
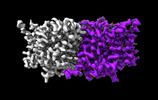
EMDB-29883: 
CLC-ec1 E202Y at pH 4.5 100mM Cl Turn
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A
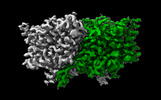
EMDB-29884: 
CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Swap
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A
EMDB-29885: 
CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Turn
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A
EMDB-29890: 
CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Intermediate
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A
EMDB-29899: 
CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Twist
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A

PDB-8ga0: 
CLC-ec1 E202Y at pH 4.5 100mM Cl Turn
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A

PDB-8ga1: 
CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Swap
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A

PDB-8ga3: 
CLC-ec1 R230C/L249C/C85A at pH 4.5 100mM Cl Turn
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A

PDB-8ga5: 
CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Intermediate
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A

PDB-8gah: 
CLC-ec1 L25C/A450C/C85A at pH 4.5 100mM Cl Twist
Method: single particle / : Fortea E, Lee S, Argyos Y, Chadda R, Ciftci D, Huysmans G, Robertson JL, Boudker O, Accardi A

EMDB-41946: 
Firmicutes Rubisco
Method: single particle / : Kaeser BP, Liu AK, Shih PM

EMDB-29725: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

EMDB-29731: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

PDB-8g4m: 
Vaccine-elicited human antibody 2C06 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Morano NC, Shapiro L, Kwong PD

PDB-8g4t: 
Vaccine-elicited human antibody 2C09 in complex with HIV-1 envelope trimer BG505 DS-SOSIP
Method: single particle / : Wang S, Kwong PD

EMDB-26313: 
C6-guano bound Mu Opioid Receptor-Gi Protein Complex
Method: single particle / : Wang H, Kobilka B

PDB-7u2k: 
C6-guano bound Mu Opioid Receptor-Gi Protein Complex
Method: single particle / : Wang H, Kobilka B

EMDB-25401: 
5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

EMDB-25402: 
5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Cao C, Roth BL, Skiniotis G

EMDB-25403: 
5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

PDB-7srq: 
5-HT2B receptor bound to LSD obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

PDB-7srr: 
5-HT2B receptor bound to LSD in complex with heterotrimeric mini-Gq protein obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Cao C, Roth BL, Skiniotis G

PDB-7srs: 
5-HT2B receptor bound to LSD in complex with beta-arrestin1 obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Cao C, Panova O, Roth BL, Skiniotis G

EMDB-26314: 
C5guano-uOR-Gi-scFv16
Method: single particle / : Wang H, Qu Q, Skiniotis G, Kobilka B

PDB-7u2l: 
C5guano-uOR-Gi-scFv16
Method: single particle / : Wang H, Qu Q, Skiniotis G, Kobilka B

EMDB-25076: 
LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

EMDB-25077: 
GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

PDB-7sf7: 
LPHN3 (ADGRL3) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

PDB-7sf8: 
GPR56 (ADGRG1) 7TM domain bound to tethered agonist in complex with G protein heterotrimer
Method: single particle / : Barros-Alvarez X, Panova O, Skiniotis G

EMDB-24733: 
Oxytocin receptor (OTR) bound to oxytocin in complex with a heterotrimeric Gq protein
Method: single particle / : Meyerowitz JG, Panova O, Skiniotis G

PDB-7ryc: 
Oxytocin receptor (OTR) bound to oxytocin in complex with a heterotrimeric Gq protein
Method: single particle / : Meyerowitz JG, Robertson MJ, Skiniotis G

EMDB-25043: 
Nuc147 bound to multiple BRCTs
Method: single particle / : Muthurajan UM, Rudolph J

PDB-7scz: 
Nuc147 bound to multiple BRCTs
Method: single particle / : Muthurajan UM, Rudolph J

EMDB-25042: 
Nuc147 bound to single BRCT
Method: single particle / : Muthurajan UM, Rudolph JR

PDB-7scy: 
Nuc147 bound to single BRCT
Method: single particle / : Muthurajan UM, Rudolph JR

EMDB-12453: 
Cryo-EM structure of the Lin28B nucleosome core particle
Method: single particle / : Roberts GA, Ozkan B, Gachulincova I, O Dwyer MR, Hall-Ponsele E, Saxena M, Robinson PJ, Soufi A

PDB-7nl0: 
Cryo-EM structure of the Lin28B nucleosome core particle
Method: single particle / : Roberts GA, Ozkan B, Gachulincova I, O Dwyer MR, Hall-Ponsele E, Saxena M, Robinson PJ, Soufi A
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
