-Search query
-Search result
Showing all 34 items for (author: robert & j & ford)

EMDB-28540: 
BG505 UFO-E2p-L4P nanoparticle reconstructed by focused refinement with a mask around the nanoparticle core
Method: single particle / : Antanasijevic A, Zhang YN, Zhu J, Ward AB
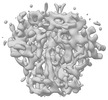
EMDB-28541: 
BG505 UFO trimer map reconstructed from BG505 UFO-E2p-L4P nanoparticle by localized reconstruction
Method: single particle / : Antanasijevic A, Zhang YN, Zhu J, Ward AB

EMDB-28542: 
BG505 UFO-10GS-I3-01v9-L7P nanoparticle reconstructed by focused refinement with a mask around the nanoparticle core
Method: single particle / : Antanasijevic A, Zhang YN, Zhu J, Ward AB

EMDB-28543: 
BG505 UFO trimer map reconstructed from BG505 UFO-10GS-I3-01v9-L7P nanoparticle by localized reconstruction
Method: single particle / : Antanasijevic A, Zhang YN, Zhu J, Ward AB

PDB-8eqn: 
BG505 UFO-E2p-L4P nanoparticle reconstructed by focused refinement with a mask around the nanoparticle core
Method: single particle / : Antanasijevic A, Zhang YN, Zhu J, Ward AB

EMDB-28596: 
CryoEM Structure of NLRP3 NACHT domain in complex with G2394
Method: single particle / : Murray JM, Johnson MC

PDB-8etr: 
CryoEM Structure of NLRP3 NACHT domain in complex with G2394
Method: single particle / : Murray JM, Johnson MC

EMDB-22627: 
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2082
Method: single particle / : Binshtein E, Crowe JE

EMDB-22628: 
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2479
Method: single particle / : Binshtein E, Crowe JE

EMDB-22148: 
Negative stain EM map of SARS-COV-2 spike protein open RBD (trimer) with Fab COV2-2096
Method: single particle / : Binshtein E, Crowe JE

EMDB-22149: 
Negative stain EM map of SARS-COV-2 spike protein (trimer) with Fab COV2-2832
Method: single particle / : Binshtein E, Crowe JE

EMDB-22094: 
CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942
Method: single particle / : LaFrance BJ, Nichols RJ, Phillips NR, Oltrogge LM, Valentin-Alvarado LE, Bischoff AJ, Savage DF, Nogales E

EMDB-22095: 
CryoEM structure of the apo-SrpI encapasulin complex from Synechococcus elongatus PCC 7942
Method: single particle / : LaFrance BJ, Nichols RJ, Phillips NR, Oltrogge LM, Valentin-Alvarado LE, Bischoff AJ, Savage DF, Nogales E

PDB-6x8m: 
CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942
Method: single particle / : LaFrance BJ, Nichols RJ, Phillips NR, Oltrogge LM, Valentin-Alvarado LE, Bischoff AJ, Savage DF, Nogales E

PDB-6x8t: 
CryoEM structure of the apo-SrpI encapasulin complex from Synechococcus elongatus PCC 7942
Method: single particle / : LaFrance BJ, Nichols RJ, Phillips NR, Oltrogge LM, Valentin-Alvarado LE, Bischoff AJ, Savage DF, Nogales E

EMDB-0021: 
Single protofilament beta-2-microglobulin amyloid fibril
Method: helical / : Iadanza MG, Ranson NA

EMDB-9363: 
Structure of M. spretus Endogenous Virus Element (EVE) Virus-like particle (VLP)
Method: single particle / : Callaway HM, Subramanian S

PDB-6nf9: 
Structure of M. spretus Endogenous Virus Element (EVE) Virus-like particle (VLP)
Method: single particle / : Callaway HM, Subramanian S

EMDB-0014: 
Two protofilament beta-2-microglobulin amyloid fibril
Method: helical / : Iadanza MG, Ranson NA

PDB-6gk3: 
Two protofilament beta-2-microglobulin amyloid fibril
Method: helical / : Iadanza MG, Ranson NA

EMDB-3611: 
Full-length dodecameric S. typhimurium Wzz complex with associated dodecyl maltoside micelle.
Method: single particle / : Ford RC, Kargas V, Collins RF, Whitfield C, Clarke BR, Siebert A, Bond PJ, Clare DK

PDB-5nbz: 
Wzz dodecamer fitted by MDFF to the Wzz experimental map from cryo-EM
Method: single particle / : Ford RC, Kargas V, Collins RF, Whitfield C, Clarke BR, Siebert A, Bond PJ, Clare DK

EMDB-8361: 
Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit
Method: single particle / : Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford Jr JL, Tong L, Frank J

PDB-5t5h: 
Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit
Method: single particle / : Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford Jr JL, Tong L, Frank J

EMDB-2626: 
The Cryo-Electron Microscopy Structure of the CorA channel from Methanocaldococcus jannaschii at 21.6 Angstrom in low magnesium.
Method: single particle / : Cleverley RM, Kean J, Shintre CA, Baldock C, Derrick JP, Ford RC, Prince SM

PDB-4cy4: 
The Cryo-Electron Microscopy Structure of the CorA channel from Methanocaldococcus jannaschii at 21.6 Angstrom in low magnesium.
Method: single particle / : Cleverley RM, Kean J, Shintre CA, Baldock C, Derrick JP, Ford RC, Prince SM

PDB-4av2: 
Single particle electron microscopy of PilQ dodecameric complexes from Neisseria meningitidis.
Method: single particle / : Berry JL, Phelan MM, Collins RF, Adomavicius T, Tonjum T, Frye SA, Bird L, Owens R, Ford RC, Lian LY, Derrick JP

EMDB-2105: 
Single particle electron microscopy of PilQ dodecameric complexes from Neisseria meningitidis.
Method: single particle / : Berry JL, Phelan MM, Collins RF, Adomavicius T, Tonjum T, Frye S, Bird L, Owens R, Ford RC, Lian LY, Derrick JP

PDB-3zx8: 
Cryo-EM reconstruction of native and expanded Turnip Crinkle virus
Method: single particle / : Bakker SE, Robottom J, Hogle JM, Maeda A, Pearson AR, Stockley PG, Ranson NA, Harrison SC

PDB-3zx9: 
Cryo-EM reconstruction of native and expanded Turnip Crinkle virus
Method: single particle / : Bakker SE, Robottom J, Pearson AR, Stockley PG, Ranson NA

EMDB-1863: 
Cryo-EM reconstruction of native and expanded Turnip Crinkle virus
Method: single particle / : Bakker SE, Robottom J, Pearson AR, Stockley PG, Ranson NA

EMDB-1864: 
Cryo-EM reconstruction of native and expanded Turnip Crinkle virus
Method: single particle / : Bakker SE, Robottom J, Pearson AR, Stockley PG, Ranson NA

PDB-4a82: 
Fitted model of staphylococcus aureus sav1866 model ABC transporter in the human cystic fibrosis transmembrane conductance regulator volume map EMD-1966.
Method: electron crystallography / : Rosenberg MF, ORyan LP, Hughes G, Zhao Z, Aleksandrov LA, Riordan JR, Ford RC

EMDB-1966: 
CFTR map generated from 2D crystals grown using the epitaxial method.
Method: electron crystallography / : Rosenberg MF, ORyan LP, Hughes G, Zhao Z, Aleksandrov LA, Riordan JR, Ford RC
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
