-Search query
-Search result
Showing all 40 items for (author: rhodes & d)
EMDB-31806: 
Telomeric mononucleosome
Method: single particle / : Soman A
EMDB-31810: 
Telomeric Dinucleosome
Method: single particle / : Soman A
EMDB-31815: 
Telomeric Dinucleosome in open state
Method: single particle / : Soman A
EMDB-31816: 
Telomeric trinucleosome
Method: single particle / : Soman A

EMDB-31823: 
Telomeric tetranucleosome
Method: single particle / : Soman A
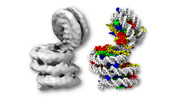
EMDB-31826: 
Telomeric trinucleosome in open state
Method: single particle / : Soman A

EMDB-31832: 
Telomeric tetranucleosome in open state
Method: single particle / : Soman A

EMDB-31907: 
Telomeric Dinucleosome at 4.6 angstroms
Method: single particle / : Soman A
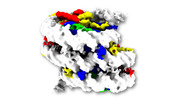
EMDB-31908: 
Telomeric Dinucleosome at 5angstroms
Method: single particle / : Soman A
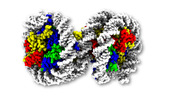
EMDB-31909: 
Open dinucleosome at 6.6 angstroms
Method: single particle / : Soman A

EMDB-13785: 
Cryo-EM map of clamped S.cerevisiae condensin-DNA complex (pentamer dataset)
Method: single particle / : Lee BG, Rhodes J, Lowe J

EMDB-13786: 
Cryo-EM structure of S.cerevisiae condensin Ycg1-Brn1-DNA complex
Method: single particle / : Lee BG, Rhodes J, Lowe J

EMDB-13787: 
Cryo-EM map of clamped S.cerevisiae condensin complex on circular DNA
Method: single particle / : Lee BG, Rhodes J, Lowe J

EMDB-13788: 
Cryo-EM map of S.cerevisiae condensin Ycg1-Brn1 complex on circular DNA
Method: single particle / : Lee BG, Rhodes J, Lowe J

PDB-7q2z: 
Cryo-EM structure of S.cerevisiae condensin Ycg1-Brn1-DNA complex
Method: single particle / : Lee BG, Rhodes J, Lowe J

EMDB-13783: 
Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (Form I)
Method: single particle / : Lee BG, Rhodes J, Lowe J

EMDB-13784: 
Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (form II)
Method: single particle / : Lee BG, Rhodes J, Lowe J

PDB-7q2x: 
Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (Form I)
Method: single particle / : Lee BG, Rhodes J, Lowe J

PDB-7q2y: 
Cryo-EM structure of clamped S.cerevisiae condensin-DNA complex (form II)
Method: single particle / : Lee BG, Rhodes J, Lowe J

EMDB-30596: 
Cryo-EM map of POT1 bound by TPP1 in closed conformation
Method: single particle / : Smith EW, Liu ZB, Lattmann S, Ahsan B, Rhodes D

EMDB-30597: 
Cryo-EM map of POT1 bound by TPP1 in extended conformation
Method: single particle / : Smith EW, Liu ZB, Lattmann S, Ahsan B, Rhodes D

EMDB-4704: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

EMDB-4705: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

EMDB-4710: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

EMDB-4711: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 4
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

EMDB-4712: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 5
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

PDB-6r1t: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

PDB-6r1u: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

PDB-6r25: 
Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3
Method: single particle / : Marabelli C, Pilotto S, Chittori S, Subramaniam S, Mattevi A

EMDB-6291: 
16 Angstrom cryo-EM reconstruction of the alpha, beta, gamma TF55 chaperonin
Method: single particle / : Chaston JJ, Stewart AG, Smits C, Aragao D, Struwe W, Benesch J, Xwong A, Ling M, Ashsan B, Sandin S, Rhodes D, Molugu SK, Bernal RA, Stock D

EMDB-2310: 
Three-dimensional structure of active, full-length human telomerase dimer, determined by single-particle electron microscopy in negative stain
Method: single particle / : Sauerwald A, Sandin S, Cristofari G, Scheres SHW, Lingner J, Rhodes D

EMDB-2311: 
Three-dimensional structure of active, full-length human telomerase. Independently refined open monomer structure, determined by single-particle electron microscopy in negative stain
Method: single particle / : Sauerwald A, Sandin S, Cristofari G, Scheres SHW, Lingner J, Rhodes D

EMDB-2312: 
Three-dimensional structure of active, full-length human telomerase. Independently refined closed monomer structure, determined by single-particle electron microscopy in negative stain
Method: single particle / : Sauerwald A, Sandin S, Cristofari G, Scheres SHW, Lingner J, Rhodes D
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search










 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
