-Search query
-Search result
Showing 1 - 50 of 338 items for (author: reddy & v)

EMDB-43629: 
Cryo-EM structure of phage DEV ejection proteins gp72:gp73
Method: single particle / : Iglesias SM, Cingolani G

PDB-8vxq: 
Cryo-EM structure of phage DEV ejection proteins gp72:gp73
Method: single particle / : Iglesias SM, Cingolani G
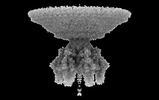
EMDB-41649: 
P22 Mature Virion tail - C6 Localized Reconstruction
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

EMDB-41651: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

EMDB-41819: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

PDB-8tvr: 
In situ cryo-EM structure of bacteriophage P22 tail hub protein: tailspike protein complex at 2.8A resolution
Method: single particle / : Iglesias S, Cingolani G, Feng-Hou C

PDB-8tvu: 
In situ cryo-EM structure of bacteriophage P22 portal protein: head-to-tail protein complex at 3.0A resolution
Method: single particle / : Iglesias SM, Cingolani G, Feng-Hou C

PDB-8u1o: 
In situ cryo-EM structure of bacteriophage P22 tailspike protein complex at 3.4A resolution
Method: single particle / : Iglesias SM, Feng-Hou C, Cingolani G

EMDB-41791: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

EMDB-41792: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

PDB-8u10: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

PDB-8u11: 
In situ cryo-EM structure of bacteriophage P22 gp1:gp5:gp4: gp10: gp9 N-term complex in conformation 2 at 3.1A resolution
Method: single particle / : Iglesias S, Feng-Hou C, Cingolani G

EMDB-28280: 
In situ cryo-EM structure of Pseudomonas phage E217 tail baseplate in C6 map
Method: single particle / : Li F, Cingolani G, Hou C

EMDB-28405: 
Pseudomonas phage E217 baseplate complex
Method: single particle / : Li F, Cingolani G, Hou C

EMDB-29406: 
Pseudomonas phage E217 5-fold vertex (capsid and decorating proteins)
Method: single particle / : Li F, Cingolani G, Hou C

EMDB-29481: 
Pseudomonas phage E217 extended sheath and tail tube
Method: single particle / : Li F, Cingolani G, Hou C

EMDB-29486: 
Pseudomonas phage E217 contracted sheath
Method: single particle / : Li F, Cingolani G, Hou C

EMDB-29487: 
Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins)
Method: single particle / : Li F, Cingolani G, Hou C

PDB-8env: 
In situ cryo-EM structure of Pseudomonas phage E217 tail baseplate in C6 map
Method: single particle / : Li F, Cingolani G, Hou C

PDB-8eon: 
Pseudomonas phage E217 baseplate complex
Method: single particle / : Li F, Cingolani G, Hou C

PDB-8frs: 
Pseudomonas phage E217 5-fold vertex (capsid and decorating proteins)
Method: single particle / : Li F, Cingolani G, Hou C

PDB-8fuv: 
Pseudomonas phage E217 extended sheath and tail tube
Method: single particle / : Li F, Cingolani G, Hou C

PDB-8fvg: 
Pseudomonas phage E217 contracted sheath
Method: single particle / : Li F, Cingolani G, Hou C

PDB-8fvh: 
Pseudomonas phage E217 neck (portal, head-to-tail connector, collar and gateway proteins)
Method: single particle / : Li F, Cingolani G, Hou C

EMDB-29542: 
4 degree_iOFS
Method: single particle / : Huang Y, Boudker O

EMDB-29543: 
4 degree_IFS-B1;
Method: single particle / : Huang Y, Boudker O

EMDB-29544: 
4 degree_IFS-A1
Method: single particle / : Huang Y, Boudker O

EMDB-29545: 
4 degree_IFS-A2
Method: single particle / : Huang Y, Boudker O

EMDB-29029: 
Langya Virus Fusion Protein (LayV-F) in Pre-Fusion Conformation
Method: single particle / : May AJ, Pothula KR, Janowska K, Acharya P

EMDB-29032: 
Langya Virus Fusion Protein (LayV-F) in Post-Fusion Conformation
Method: single particle / : May AJ, Pothula KR, Janowska K, Acharya P

PDB-8fej: 
Langya Virus Fusion Protein (LayV-F) in Pre-Fusion Conformation
Method: single particle / : May AJ, Pothula KR, Janowska K, Acharya P

PDB-8fel: 
Langya Virus Fusion Protein (LayV-F) in Post-Fusion Conformation
Method: single particle / : May AJ, Pothula KR, Janowska K, Acharya P

EMDB-26656: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32
Method: single particle / : Patel A, Ortlund E

PDB-7uow: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32
Method: single particle / : Patel A, Ortlund E

EMDB-26497: 
Asp-bound GltPh RSMR mutant in IFS-A2 state
Method: single particle / : Huang Y, Boudker O

EMDB-26498: 
Asp-bound GltPh RSMR mutant in IFS-B1 state
Method: single particle / : Huang Y, Boudker O

EMDB-26263: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-02
Method: single particle / : Patel A, Ortlund E

EMDB-26267: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-13
Method: single particle / : Patel A, Ortlund E

PDB-7u0q: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-02
Method: single particle / : Patel A, Ortlund E

PDB-7u0x: 
SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-13
Method: single particle / : Patel A, Ortlund E
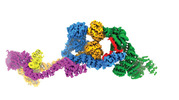
EMDB-29073: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J
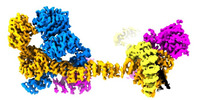
EMDB-29078: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J

PDB-8fgw: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J

PDB-8fh3: 
Human IFT-A complex structures provide molecular insights into ciliary transport
Method: single particle / : Jiang M, Palicharla VR, Miller D, Hwang SH, Zhu H, Hixson P, Mukhopadhyay S, Sun J

EMDB-27337: 
Open MscS in PC14.1 Nanodiscs
Method: single particle / : Reddy BG, Bavi N, Perozo E

EMDB-26065: 
nsEM maps of coxsackievirus A21 viral particle alone and in complex with mouse polyclonal antibodies
Method: single particle / : Antanasijevic A, Ward AB

EMDB-26068: 
Coxsackievirus A21 capsid subdomain in complex with mouse polyclonal antibody pAbC-3
Method: single particle / : Antanasijevic A, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
