-Search query
-Search result
Showing all 36 items for (author: reck-peterson & sl)

EMDB-27813: 
Structure of monomeric LRRK1
Method: single particle / : Reimer JM, Mathea S, Chatterjee D, Knapp S, Leschziner AE

EMDB-27814: 
Local refinement around RCKW of LRRK1
Method: single particle / : Reimer JM, Mathea S, Knapp S, Leschziner AE

EMDB-27815: 
Local refinement of LRRK1 around the ROC-COR-kinase domains
Method: single particle / : Reimer JM, Mathea S, Knapp S, Leschziner AE

EMDB-27816: 
Local refinement around kinase and WD40 domains of LRRK1
Method: single particle / : Reimer JM, Mathea S, Knapp S, Leschziner AE

EMDB-27817: 
Structure of dimeric LRRK1
Method: single particle / : Reimer JM, Lin YX, Leschziner AE

EMDB-27818: 
Symmetry expansion of dimeric LRRK1
Method: single particle / : Reimer JM, Lin YX, Leschziner AE

EMDB-27810: 
Cryo-EM structure of chi dynein bound to Lis1
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-27811: 
Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation.
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-27782: 
Structure of human cytoplasmic dynein-1 bound to two Lis1 proteins
Method: single particle / : Reimer JM, DeSantis M, Reck-Peterson SL, Leschziner AE

EMDB-27783: 
Structure of human cytoplasmic dynein-1 bound to one Lis1
Method: single particle / : Reimer JM, DeSantis M, Reck-Peterson SL, Leschziner AE

EMDB-25649: 
Helical Reconstruction of the C-terminal Half of Leucine Rich Repeat Kinase 2 (LRRK2) (I2020T) bound to 11-protofilament microtubule in presence of MLi-2 kinase inhibitor
Method: helical / : Matyszewski M, Leschziner AE

EMDB-25658: 
Local refinement of the C-terminal Half of Leucine Rich Repeat Kinase 2 (LRRK2) (I2020T) bound to 11-protofilament microtubule in presence of MLi-2 kinase inhibitor
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-25664: 
Local Refinement of the C-terminal Half of Leucine Rich Repeat Kinase 2 (LRRK2) (I2020T) tetramer bound to 11-protofilament microtubule in presence of MLi-2 kinase inhibitor
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-25672: 
Structure of the C-terminal half of Leucine Rich Repeat Kinase 1 (LRRK1)
Method: single particle / : Matyszewski M, Snead DM, Leschziner AE

EMDB-25674: 
Local refinement of the C-terminal Half of Leucine Rich Repeat Kinase 2 (LRRK2) (I2020T) bound to 12-protofilament microtubule without kinase inhibitors present
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-25897: 
Local refinement of the C-terminal Half of Leucine Rich Repeat Kinase 2 (LRRK2) (I2020T) bound to 11-protofilament microtubule focused on the monomer in presence of MLi-2 kinase inhibitor
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-25906: 
Structure of Leucine Rich Repeat Kinase 2's ROC domain interacting with the microtubule facing the minus end
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-25907: 
Structure of Leucine Rich Repeat Kinase 2's ROC domain interacting with the microtubule facing the plus end
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-25908: 
2 microtubule protofilaments locally refined from Leucine Rich Repeat Kinase 2 filament structure bound to 11-protofilament microtubules
Method: single particle / : Matyszewski M, Leschziner AE

EMDB-23829: 
Structure of yeast cytoplasmic dynein with AAA3 Walker B mutation bound to Lis1
Method: single particle / : Lahiri I, Reimer JM, Leschziner AE

EMDB-25497: 
Signal-subtracted reconstruction of yeast cytoplasmic dynein-1 (AAA3 Walker B mutant, E2488Q) AAA2-AAA5L domains bound to Lis1
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-21250: 
Cryo-EM structure of the C-terminal half of the Parkinson's Disease-linked protein Leucine Rich Repeat Kinase 2 (LRRK2)
Method: single particle / : Leschziner A, Deniston C, Lahiri I

EMDB-21306: 
Cryo-EM map of the C-terminal half of Leucine Rich Repeat Kinase 2 as a monomer at 8.1 angstroms
Method: single particle / : Leschziner A, Deniston C, Lahiri I

EMDB-21309: 
Cryo-EM map of C-terminal half of Leucine Rich Repeat Kinase 2 as a COR-COR domain dimer at 9.5 angstroms
Method: single particle / : Leschziner A, Deniston C, Lahiri I

EMDB-21310: 
Cryo-EM map of C-terminal half of Leucine Rich Repeat Kinase 2 as a WD40-WD40 domain dimer at 13.4 angstroms
Method: single particle / : Leschziner A, Deniston C, Lahiri I

EMDB-21311: 
Cryo-EM map of C-terminal half of Leucine Rich Repeat Kinase 2 as a COR-COR domain dimer in the presence of the MLi-2 kinase inhibitor at 9.0 angstroms
Method: single particle / : Leschziner A, Deniston C, Lahiri I

EMDB-21312: 
Cryo-EM map of C-terminal half of Leucine Rich Repeat Kinase 2 as a WD40-WD40 domain dimer in the presence of the MLi-2 kinase inhibitor at 10.2 angstroms
Method: single particle / : Leschziner A, Deniston C, Lahiri I

EMDB-8673: 
Cryo-EM structure of yeast cytoplasmic dynein-1 with Lis1 and ATP
Method: single particle / : Cianfrocco MA, DeSantis ME, Htet ZM, Tran PT, Reck-Peterson SL, Leschziner AE

EMDB-8706: 
Cryo-EM structure of yeast cytoplasmic dynein with Walker B mutation at AAA3 in presence of ATP-VO4
Method: single particle / : Cianfrocco MA, DeSantis ME, Htet ZM, Tran PT, Reck-Peterson SL, Leschziner AE

EMDB-6008: 
Dynein motor domain in complex with Lis1 in the absence of nucleotide
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE
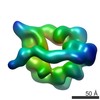
EMDB-6013: 
Dynein motor domain in the absence of nucleotide
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE

EMDB-6014: 
Dynein motor domain in the presence of ADP, with linker at position 2
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE
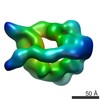
EMDB-6015: 
Dynein motor domain in the presence of ADP, with linker at position 1
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE
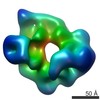
EMDB-6016: 
Dynein motor domain in complex with Lis1 in the presence of ATP and Vi
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE

EMDB-6017: 
Dynein motor domain with truncated linker in complex with Lis1 in the absence of nucleotide
Method: single particle / : Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, Reck-Peterson SL, Leschziner AE

EMDB-5439: 
Structural basis for microtubule binding and release by dynein
Method: helical / : Redwine WB, Hernandez-Lopez R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
