-Search query
-Search result
Showing 1 - 50 of 83 items for (author: rasmussen & t)

EMDB-28669: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 01)
Method: electron tomography / : Liu J, Ren G
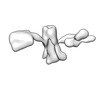
EMDB-28670: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 02)
Method: electron tomography / : Liu J, Ren G
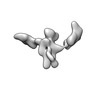
EMDB-28671: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 03)
Method: electron tomography / : Liu J, Ren G
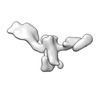
EMDB-28672: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 04)
Method: electron tomography / : Liu J, Ren G

EMDB-28673: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 05)
Method: electron tomography / : Liu J, Ren G
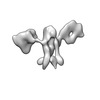
EMDB-28674: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 06)
Method: electron tomography / : Liu J, Ren G
EMDB-28675: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 07)
Method: electron tomography / : Liu J, Ren G
EMDB-28676: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 08)
Method: electron tomography / : Liu J, Ren G

EMDB-28677: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 09)
Method: electron tomography / : Liu J, Ren G

EMDB-28678: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 10)
Method: electron tomography / : Liu J, Ren G

EMDB-28679: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 11)
Method: electron tomography / : Liu J, Ren G

EMDB-28680: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 12)
Method: electron tomography / : Liu J, Ren G

EMDB-28681: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 13)
Method: electron tomography / : Liu J, Ren G

EMDB-28682: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 14)
Method: electron tomography / : Liu J, Ren G
EMDB-28683: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 15)
Method: electron tomography / : Liu J, Ren G
EMDB-28684: 
Single-Molecule 3D Image of 16 Helix RNA Origami Satellite by Individual Particle Electron Tomography (No. 16)
Method: electron tomography / : Liu J, Ren G

EMDB-13926: 
Twist-corrected RNA origami 5-helix Tile A
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

PDB-7qdu: 
Twist-corrected RNA origami 5-helix Tile A
Method: single particle / : McRae EKS, Andersen ES

EMDB-13625: 
Mature conformer 2 of a 6-helix bundle of RNA with a clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13626: 
Young conformer 2 of a 6-helix bundle of RNA with a clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13627: 
6-Helix bundle of RNA
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13628: 
Young conformer of a 6-helix bundle of RNA with clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13630: 
Mature conformer of a 6-helix bundle of RNA with clasp
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13633: 
RNA origami 5-helix tile
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13636: 
RNA origami 5-helix tile
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

PDB-7ptk: 
Young conformer of a 6-helix bundle of RNA with clasp
Method: single particle / : McRae EKS, Andersen ES

PDB-7ptl: 
Mature conformer of a 6-helix bundle of RNA with clasp
Method: single particle / : McRae EKS, Andersen ES

PDB-7ptq: 
RNA origami 5-helix tile
Method: single particle / : McRae EKS, Andersen ES

PDB-7pts: 
RNA origami 5-helix tile
Method: single particle / : McRae EKS, Andersen ES

EMDB-13592: 
Cryo-EM SPA reconstruction of an extended RNA origami 5 helix tile (5HT-B-3X)
Method: single particle / : McRae EKS, Bogglid A, Boesen T, Andersen ES

EMDB-13726: 
HBc-F97L premature secretion phenotype
Method: single particle / : Makbul C, Boettcher B

EMDB-13728: 
HBc-F97L (premature secretion phenotype) in complex with Triton X-100
Method: single particle / : Makbul C, Boettcher B

EMDB-13731: 
HBc-WT in complex with Triton X-100
Method: single particle / : Makbul C, Boettcher B

EMDB-13732: 
HBc-F97L premature secretion phenotype
Method: single particle / : Makbul C, Boettcher B

EMDB-13733: 
HBc-P5T in complex with X-100
Method: single particle / : Makbul C, Boettcher B

EMDB-13734: 
wt HBc capsid like particles in complex with inhibitory peptide SLLGRM and Triton X-100
Method: single particle / : Makbul C, Boettcher B

PDB-7pz9: 
HBc-F97L premature secretion phenotype
Method: single particle / : Makbul C, Boettcher B

PDB-7pzi: 
HBc-F97L (premature secretion phenotype) in complex with Triton X-100
Method: single particle / : Makbul C, Boettcher B

PDB-7pzk: 
HBc-WT in complex with Triton X-100
Method: single particle / : Makbul C, Boettcher B

PDB-7pzl: 
HBc-F97L premature secretion phenotype
Method: single particle / : Makbul C, Boettcher B

PDB-7pzm: 
HBc-P5T in complex with X-100
Method: single particle / : Makbul C, Boettcher B

PDB-7pzn: 
wt HBc capsid like particles in complex with inhibitory peptide SLLGRM and Triton X-100
Method: single particle / : Makbul C, Boettcher B

EMDB-13048: 
cytochrome bd-II type oxidase with bound aurachin D
Method: single particle / : Grauel A, Kaegi J

PDB-7ose: 
cytochrome bd-II type oxidase with bound aurachin D
Method: single particle / : Grauel A, Kaegi J, Rasmussen T, Wohlwend D, Boettcher B, Friedrich T

EMDB-12996: 
Mechanosensitive channel MscS solubilized with LMNG in open conformation
Method: single particle / : Rasmussen T, Flegler VJ, Boettcher B

EMDB-12997: 
Mechanosensitive channel MscS solubilized with DDM in closed conformation
Method: single particle / : Rasmussen T, Flegler VJ, Boettcher B

EMDB-13003: 
Mechanosensitive channel MscS solubilized with DDM in open conformation
Method: single particle / : Rasmussen T, Flegler VJ, Boettcher B

EMDB-13006: 
Mechanosensitive channel MscS solubilized with DDM in closed conformation with added lipid
Method: single particle / : Rasmussen T, Flegler VJ, Boettcher B

EMDB-13007: 
Mechanosensitive channel MscS solubilized with LMNG in closed conformation with added lipid
Method: single particle / : Rasmussen T, Flegler VJ, Boettcher B

EMDB-13008: 
Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid
Method: single particle / : Rasmussen T, Flegler VJ, Boettcher B
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
