-Search query
-Search result
Showing all 47 items for (author: porta & c)

EMDB-28845: 
Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

EMDB-29640: 
Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

EMDB-29676: 
Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

EMDB-29683: 
Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

EMDB-29732: 
Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

EMDB-29812: 
Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

EMDB-29859: 
Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand
Method: single particle / : Porta JC, Ohi MD, Walter NG, Frank AT, Deb I, Meze K

PDB-8f3c: 
Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

PDB-8g00: 
Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

PDB-8g1s: 
Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

PDB-8g2w: 
Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

PDB-8g4w: 
Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

PDB-8g7e: 
Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand
Method: single particle / : Porta JC, Chauvier A, Deb I, Ellinger E, Frank AT, Meze K, Ohi MD, Walter NG

PDB-8g8z: 
Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand
Method: single particle / : Porta JC, Ohi MD, Walter NG, Frank AT, Deb I, Meze K

EMDB-13795: 
Cryo-EM structure of TDP43 core peptide amyloid fiber
Method: helical / : Nazarov S, Lashuel H

EMDB-15725: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3
Method: single particle / : Bahar MW, Fry EE, Stuart DI
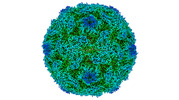
EMDB-15726: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril
Method: single particle / : Bahar MW, Fry EE, Stuart DI
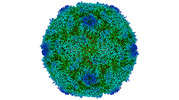
EMDB-15727: 
Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8ayx: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8ayy: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8ayz: 
Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17
Method: single particle / : Bahar MW, Fry EE, Stuart DI
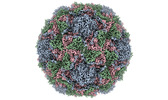
EMDB-15543: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8).
Method: single particle / : Bahar MW, Fry EE, Stuart DI

PDB-8anw: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8).
Method: single particle / : Bahar MW, Fry EE, Stuart DI

EMDB-25007: 
CryoEM structure of the Caveolin-1 8S complex
Method: single particle / : Porta JP, Ohi MD, Kenworthy AK, Karakas E

PDB-7sc0: 
CryoEM structure of the Caveolin-1 8S complex
Method: single particle / : Porta JP, Ohi MD, Kenworthy AK, Karakas E

EMDB-13054: 
Cryo-EM structure of nonameric EPEC SctV-C
Method: single particle / : Yuan B, Wald J, Fahrenkamp D, Marlovits TC

PDB-7osl: 
Cryo-EM structure of nonameric EPEC SctV-C
Method: single particle / : Fahrenkamp D, Wald J, Yuan B, Marlovits TC

EMDB-11106: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) from a mammalian expression system.
Method: single particle / : Bahar MW, Porta C, Fry EE, Stuart DI

PDB-6z6w: 
Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) from a mammalian expression system.
Method: single particle / : Bahar MW, Porta C, Fry EE, Stuart DI

EMDB-8548: 
A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection
Method: single particle / : Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS, Crowe Jr JE, Kuhn RJ, Rossmann MG

PDB-5uhy: 
A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection
Method: single particle / : Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS, Crowe Jr JE, Kuhn RJ, Rossmann MG

EMDB-3148: 
Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab 4J21
Method: single particle / : Long F, Fong RH, Austin SK, Chen Z, Klose T, Fokine A, Liu Y, Porta J, Sapparapu G, Akahata W, Doranz BJ, Crowe JE, Diamond MS, Rossmann MG

EMDB-3149: 
Electron cryo-microscopy of chikungunya virus in complex with neutralizing antibody Fab 5M16
Method: single particle / : Long F, Fong RH, Austin SK, Chen Z, Klose T, Fokine A, Liu Y, Porta J, Sapparapu G, Akahata W, Doranz BJ, Crowe JE, Diamond MS, Rossmann MG

EMDB-6356: 
Electron cryo-microscopy of Chikungunya virus-like particles in complex with neutralizing antibody Fab 5F10
Method: single particle / : Porta JC, Mangala Prasad V, Akahata W, Wang CI, Ng L, Rossmann MG

EMDB-6368: 
Electron Cryo-microscopy of Chikungunya virus-like particles in complex with neutralizing antibody Fab 8B10
Method: single particle / : Porta JC, Mangala Prasad V, Wang CI, Akahata W, Ng LFP, Rossmann MG

EMDB-6394: 
Structure of Ljungan virus: insight into picornavirus packaging
Method: single particle / : Zhu L, Wang XX, Ren JS, Porta C, Wenham H, Ekstrom JO, Panjwani A, Knowles NJ, Kotecha A, Siebert A, Lindberg M, Fry EE, Rao ZH, Tuthill TJ, Stuart DI

EMDB-6395: 
Structure of Ljungan virus: insight into picornavirus packaging
Method: single particle / : Zhu L, Wang XX, Ren JS, Porta C, Wenham H, Ekstrom JO, Panjwani A, Knowles NJ, Kotecha A, Siebert A, Lindberg M, Fry EE, Rao ZH, Tuthill TJ, Stuart DI

PDB-3jb4: 
Structure of Ljungan virus: insight into picornavirus packaging
Method: single particle / : Zhu L, Wang XX, Ren JS, Porta C, Wenham H, Ekstrom JO, Panjwani A, Knowles NJ, Kotecha A, Siebert A, Lindberg M, Fry EE, Rao ZH, Tuthill TJ, Stuart DI

EMDB-3129: 
Structure-based energetics of protein interfaces guide FMDV vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, Perez-Martin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

EMDB-3130: 
Structure-based energetics of protein interfaces guide FMDV vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, Perez-Martin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

PDB-5ac9: 
Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, PerezMartin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

PDB-5aca: 
Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design
Method: single particle / : Kotecha A, Seago J, Scott K, Burman A, Loureiro S, Ren J, Porta C, Ginn HM, Jackson T, Perez-Martin E, Siebert CA, Paul G, Huiskonen JT, Jones IM, Esnouf RM, Fry EE, Maree FF, Charleston B, Stuart DI

PDB-4uom: 
Electron Cryo-microscopy of Venezuelan Equine Encephalitis Virus TC- 83 in complex with neutralizing antibody Fab F5
Method: single particle / : Porta J, Jose J, Roehrig JT, Blair CD, Kuhn RJ, Rossmann MG

PDB-4uok: 
Electron Cryo-microscopy of Venezuelan Equine Encephalitis Virus TC-83 in complex with neutralizing antibody Fab 3B4C-4
Method: single particle / : Porta J, Jose J, Roehrig JT, Blair CD, Kuhn RJ, Rossmann MG

EMDB-2645: 
Electron Cryo-microscopy of Venezuelan Equine Encephalitis Virus TC-83 in complex with neutralizing antibody Fab F5
Method: single particle / : Porta JC, Jose J, Roehrig JT, Blair CD, Kuhn RJ, Rossmann MG

EMDB-2655: 
Electron Cryo-microscopy of Venezuelan Equine Encephalitis Virus TC-83 in complex with neutralizing antibody Fab 3B4C-4
Method: single particle / : Porta JC, Jose J, Roehrig JT, Blair CD, Kuhn RJ, Rossmann MG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
