-Search query
-Search result
Showing 1 - 50 of 218 items for (author: pintilie & g)

EMDB-41637: 
Asymmetric cryoEM reconstruction of Mayaro virus
Method: single particle / : Chmielewski D, Kaelber J, Jin J, Weaver S, Auguste AJ, Chiu W

EMDB-28979: 
Cryo-EM structure of Chikungunya virus asymmetric unit
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41096: 
Cryo-electron tomography of Chikungunya virus pentamer structure
Method: subtomogram averaging / : Chmielewsk D, Su GC, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41631: 
Cryo-EM structure of Chikungunya virus with asymmetric reconstruction
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

PDB-8fcg: 
Cryo-EM structure of Chikungunya virus asymmetric unit
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W
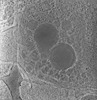
EMDB-28138: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
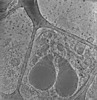
EMDB-28141: 
3D reconstruction of the apical complex of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY
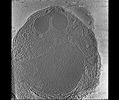
EMDB-28142: 
3D Reconstruction of Plasmodium falciparum (3D7) free merozoite
Method: electron tomography / : Segev-Zarko L, Sun SY, Kim CY

EMDB-41126: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41127: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41128: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41129: 
Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41130: 
Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

PDB-8ta2: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

PDB-8ta3: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

PDB-8ta4: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

PDB-8ta5: 
Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

PDB-8ta6: 
Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-29395: 
Subtomogram average of HuCoV-NL63 spike protein from purified intact virions
Method: subtomogram averaging / : Chen M, Chmielewski D, Schmid M, Jin J, Chiu W

PDB-8u26: 
Gaussian Mixture Models based single particle refinement - GPCR (Substance P bound to active human neurokinin 1 receptor in complex with miniGs399 from EMPIAR-10786)
Method: single particle / : Chen M, Pintilie G

PDB-8u28: 
Gaussian mixture model based single particle refinement - SARS (SARS-CoV-2 Spike Proteins on intact virions from EMPIAR-10492)
Method: single particle / : Chen M, Pintilie G

PDB-8u2c: 
Gaussian mixture model based single particle refinement - ABC transporter (inhibitor-bound ABCG2 from EMPIAR-10374)
Method: single particle / : Chen M, Pintilie G

EMDB-28125: 
Plasmodium falciparum merozoites apical 2-ring units
Method: subtomogram averaging / : Sun SY, Pintilie GD

EMDB-28126: 
Toxoplasma apical rings
Method: subtomogram averaging / : Sun SY, Pintilie GD

PDB-8fr7: 
A hinge glycan regulates spike bending and impacts coronavirus infectivity
Method: single particle / : Pintilie G, Wilson E, Chmielewski D, Schmid MF, Jin J, Chen M, Singharoy A, Chiu W

EMDB-33736: 
Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K, Liu L

EMDB-33738: 
Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K, Liu L

EMDB-33739: 
Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K, Liu L

EMDB-33740: 
Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K, Liu L

PDB-7yc8: 
Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K

PDB-7ycg: 
Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K

PDB-7ych: 
Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K

PDB-7yci: 
Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing
Method: single particle / : Zhang X, Li S, Pintilie G, Palo MZ, Zhang K

EMDB-33811: 
Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

EMDB-33812: 
Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

EMDB-33813: 
Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

EMDB-33814: 
Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

EMDB-33815: 
Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

EMDB-33816: 
Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

PDB-7yg8: 
Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

PDB-7yg9: 
Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

PDB-7yga: 
Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

PDB-7ygb: 
Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

PDB-7ygc: 
Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

PDB-7ygd: 
Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing
Method: single particle / : Li S, Michael ZP, Zhang X, Greg P, Zhang K

EMDB-32822: 
An apo TRiC map
Method: single particle / : Park JS, Roh SH

EMDB-26089: 
The beta-tubulin folding intermediate I
Method: single particle / : Zhao Y, Frydman J, Chiu W

EMDB-26120: 
The beta-tubulin folding intermediate II
Method: single particle / : Zhao Y, Frydman J, Chiu W

EMDB-26123: 
The beta-tubulin folding intermediate III
Method: single particle / : Zhao Y, Frydman J, Chiu W

EMDB-26131: 
The beta-tubulin folding intermediate IV
Method: single particle / : Zhao Y, Frydman J, Chiu W
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
