-Search query
-Search result
Showing 1 - 50 of 309 items for (author: peters & j)

EMDB-18594: 
Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs
Method: single particle / : Gao Y, Zhang Y, Hakke S, Peters PJ, Ravelli RBG

PDB-8qqk: 
Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs
Method: single particle / : Gao Y, Zhang Y, Hakke S, Peters PJ, Ravelli RBG

EMDB-19477: 
Saccharomyces cerevisiae FAS type I
Method: single particle / : Mann D, Grininger M, Ludig D, Sachse C

EMDB-19489: 
Tobacco mosaic virus from scanning transmission electron microscopy at CSA=2.0 mrad
Method: helical / : Mann D, Filopoulou A, Sachse C

EMDB-16004: 
Structure of hexameric subcomplexes (Truncation Delta2-6) of the fractal citrate synthase from Synechococcus elongatus PCC7942
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-15529: 
Structure of a first level Sierpinski triangle formed by a citrate synthase
Method: single particle / : Lo YK, Bohn S, Sendker FL, Schuller JM, Hochberg G

EMDB-16510: 
AQP7_inhibitor
Method: single particle / : Huang P, Venskutonyte R, Gourdon P, Lindkvist-Petersson K

PDB-8c9h: 
AQP7_inhibitor
Method: single particle / : Huang P, Venskutonyte R, Gourdon P, Lindkvist-Petersson K

EMDB-43137: 
SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop
Method: single particle / : Peterson JM, Becker ST, O'Leary CA, Juneja P, Yang Y, Moss WN

PDB-8vci: 
SARS-CoV-2 Frameshift Stimulatory Element with Upstream Multibranch Loop
Method: single particle / : Peterson JM, Becker ST, O'Leary CA, Juneja P, Yang Y, Moss WN

EMDB-41579: 
Structure of full-length LexA bound to a RecA filament
Method: helical / : Cory MB, Li A, Kohli RM

EMDB-27813: 
Structure of monomeric LRRK1
Method: single particle / : Reimer JM, Mathea S, Chatterjee D, Knapp S, Leschziner AE

EMDB-27814: 
Local refinement around RCKW of LRRK1
Method: single particle / : Reimer JM, Mathea S, Knapp S, Leschziner AE

EMDB-27815: 
Local refinement of LRRK1 around the ROC-COR-kinase domains
Method: single particle / : Reimer JM, Mathea S, Knapp S, Leschziner AE

EMDB-27816: 
Local refinement around kinase and WD40 domains of LRRK1
Method: single particle / : Reimer JM, Mathea S, Knapp S, Leschziner AE

EMDB-27817: 
Structure of dimeric LRRK1
Method: single particle / : Reimer JM, Lin YX, Leschziner AE

EMDB-27818: 
Symmetry expansion of dimeric LRRK1
Method: single particle / : Reimer JM, Lin YX, Leschziner AE

PDB-8e04: 
Structure of monomeric LRRK1
Method: single particle / : Reimer JM, Mathea S, Chatterjee D, Knapp S, Leschziner AE

PDB-8e05: 
Structure of dimeric LRRK1
Method: single particle / : Reimer JM, Lin YX, Leschziner AE

PDB-8e06: 
Symmetry expansion of dimeric LRRK1
Method: single particle / : Reimer JM, Lin YX, Leschziner AE

EMDB-27810: 
Cryo-EM structure of chi dynein bound to Lis1
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-27811: 
Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation.
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

PDB-8dzz: 
Cryo-EM structure of chi dynein bound to Lis1
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

PDB-8e00: 
Symmetry expansion of yeast cytoplasmic dynein-1 bound to Lis1 in the chi conformation.
Method: single particle / : Reimer JM, Lahiri I, Leschziner AE

EMDB-18010: 
Charging of vitreous samples in cryogenic electron microscopy mitigated by graphene - BfrB - Dataset 4 - Quantifoil 300 mesh R1.2/1.3 with Graphene - Large Beam
Method: single particle / : van schayck JP, Zhang Y, Ravelli RBG

EMDB-18028: 
Charging of vitreous samples in cryogenic electron microscopy mitigated by graphene - BfrB - Dataset 2 - Quantifoil 300 mesh R1.2/1.3 with Graphene - Small Beam
Method: single particle / : van schayck JP, Zhang Y, Ravelli RBG

EMDB-18029: 
Charging of vitreous samples in cryogenic electron microscopy mitigated by graphene - BfrB - Dataset 1 - Quantifoil 300 mesh R1.2/1.3 - Small Beam
Method: single particle / : van schayck JP, Zhang Y, Ravelli RBG

EMDB-18030: 
Charging of vitreous samples in cryogenic electron microscopy mitigated by graphene - BfrB - Dataset 3 - Quantifoil 300 mesh R1.2/1.3 - Large Beam
Method: single particle / : van schayck JP, Zhang Y, Ravelli RBG

EMDB-26348: 
I-F3b Cascade-TniQ full R-loop complex
Method: single particle / : Park JU, Mehrotra E, Kellogg EH

PDB-7u5d: 
I-F3b Cascade-TniQ full R-loop complex
Method: single particle / : Park JU, Mehrotra E, Kellogg EH

EMDB-26349: 
I-F3b Cascade-TniQ partial R-loop complex
Method: single particle / : Park JU, Mehrotra E, Kellogg EH

PDB-7u5e: 
I-F3b Cascade-TniQ partial R-loop complex
Method: single particle / : Park JU, Mehrotra E, Kellogg EH

EMDB-15853: 
Tetrameric structure of 47 N-terminally truncated human tryptophan hydroxylase 2 with dimerized regulatory domains
Method: single particle / : Zhang Z, Vedel IM, Skawinska NT, Harris P, Stark H, Peters GHJ

EMDB-16223: 
Cryo-EM structure of the catalytic domain tetramer of N-terminally truncated human tryptophan hydroxylase 2
Method: single particle / : Zhang Z, Vedel IM, Skawinska NT, Harris P, Stark H, Peters GHJ

EMDB-29657: 
Semi-synthetic CoA-alpha-Synuclein Constructs Trap N-terminal Acetyltransferase NatB for Binding Mechanism Studies
Method: single particle / : Gardner SM, Marmorstein R

PDB-8g0l: 
Semi-synthetic CoA-alpha-Synuclein Constructs Trap N-terminal Acetyltransferase NatB for Binding Mechanism Studies
Method: single particle / : Gardner SM, Marmorstein R

EMDB-29735: 
Structure of nucleosome-bound Sirtuin 6 deacetylase
Method: single particle / : Chio US, Rechiche O, Bryll AR, Zhu J, Feldman JL, Peterson CL, Tan S, Armache JP

PDB-8g57: 
Structure of nucleosome-bound Sirtuin 6 deacetylase
Method: single particle / : Chio US, Rechiche O, Bryll AR, Zhu J, Feldman JL, Peterson CL, Tan S, Armache JP

EMDB-28092: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-093
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28090: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-040
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO

EMDB-28091: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-045
Method: single particle / : Li H, Callaway H, Yu X, Shek J, Saphire EO
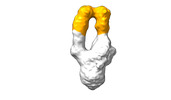
EMDB-28093: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-156
Method: single particle / : Shek J, Callaway H, Li H, Yu X, Saphire EO

EMDB-27782: 
Structure of human cytoplasmic dynein-1 bound to two Lis1 proteins
Method: single particle / : Reimer JM, DeSantis M, Reck-Peterson SL, Leschziner AE

EMDB-27783: 
Structure of human cytoplasmic dynein-1 bound to one Lis1
Method: single particle / : Reimer JM, DeSantis M, Reck-Peterson SL, Leschziner AE

PDB-8dyu: 
Structure of human cytoplasmic dynein-1 bound to two Lis1 proteins
Method: single particle / : Reimer JM, DeSantis M, Reck-Peterson SL, Leschziner AE

PDB-8dyv: 
Structure of human cytoplasmic dynein-1 bound to one Lis1
Method: single particle / : Reimer JM, DeSantis M, Reck-Peterson SL, Leschziner AE
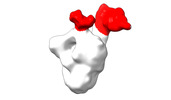
EMDB-28094: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-234
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28095: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-260
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28096: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-279
Method: single particle / : Callaway H, Li H, Yu X, Shek J, Saphire EO

EMDB-28097: 
Negative stain EM map of SARS-CoV-2 Spike in complex with CoVIC-290
Method: single particle / : Yu X, Callaway H, Li H, Shek J, Saphire EO
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
