-Search query
-Search result
Showing all 42 items for (author: paulson & jc)

EMDB-25622: 
Negative Stain EM reconstruction of ApexGT3 in complex with two copies of PG9 Fab
Method: single particle / : Berndsen ZT

EMDB-25732: 
HIV-1 Envelope ApexGT2.2MUT in complex with PCT64.LMCA Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25733: 
HIV-1 Envelope ApexGT2 in complex with PCT64.35S Fab and RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25734: 
HIV-1 Envelope ApexGT2 in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25735: 
HIV-1 Envelope ApexGT3 in complex with PG9.iGL Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-25736: 
HIV-1 Envelope ApexGT3.N130 in complex with PG9 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-22108: 
HIV-1 Envelope Glycoprotein BG505 SOSIP.664 expressed in HEK293F cells in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-22109: 
HIV-1 Envelope Glycoprotein BG505 SOSIP.664 expressed in stable CHO cells in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-22110: 
HIV-1 Envelope Glycoprotein BG505 SOSIP.664 expressed in HEK293S cells in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-22111: 
HIV-1 Envelope Glycoprotein BG505 SOSIP.664, expressed in HEK293S cells and partially deglycosylated by endoglycosidase H, in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-22112: 
HIV-1 Envelope Glycoprotein BG505 SOSIP.664, expressed in HEK293S cells and deglycosylated by endoglycosidase H, in complex with RM20A3 Fab
Method: single particle / : Berndsen ZT, Ward AB

EMDB-22352: 
Structure of SARS-CoV-2 3Q-2P full-length prefusion spike trimer (C3 symmetry)
Method: single particle / : Bangaru S, Turner HL, Ozorowski G, Antanasijevic A, Ward AB

EMDB-22353: 
Structure of SARS-CoV-2 3Q-2P full-length prefusion trimer (C1 symmetry)
Method: single particle / : Bangaru S, Turner HL, Ozorowski G, Antanasijevic A, Ward AB

EMDB-22354: 
Structure of SARS-CoV-2 3Q-2P full-length dimers of spike trimers
Method: single particle / : Bangaru S, Turner HL, Ozorowski G, Antanasijevic A, Ward AB

EMDB-22355: 
Structure of SARS-CoV-2 3Q-2P full-length trimer of spike trimers
Method: single particle / : Bangaru S, Turner HL, Ozorowski G, Antanasijevic A, Ward AB

EMDB-22356: 
Structure of SARS-CoV-2 3Q full-length prefusion trimer
Method: single particle / : Bangaru S, Turner HL, Ozorowski G, Antanasijevic A, Ward AB

EMDB-20538: 
CryoEM map of NA-22 Fab in complex with N9 Shangahi2
Method: single particle / : Ward AB, Turner HL, Zhu X

EMDB-20540: 
CryoEM map of NA-73 Fab in complex with N9 Shanghai2
Method: single particle / : Ward AB, Turner HL, Zhu X

EMDB-20541: 
CryoEM map of NA-80 Fab in complex with N9 Shangahi2
Method: single particle / : Ward AB, Turner HL, Zhu X

EMDB-20594: 
CryoEM-derived model of NA-63 Fab in complex with N9 Shanghai2
Method: single particle / : Ward AB, Turner HL

EMDB-9030: 
HIV-1 Envelope Glycoprotein Clone B41_delCT in complex with the broadly neutralizing antibody Fab PGT151
Method: single particle / : Berndsen ZT, Ward AB

EMDB-9062: 
HIV-1 Envelope Glycoprotein Clone BG505 delCT N332T in complex with broadly neutralizing antibody Fab PGT151
Method: single particle / : Berndsen ZT, Ward AB

EMDB-7858: 
Ectodomain of full length, wild type HIV-1 glycoprotein clone PC64M18C043 in complex with PGT151 Fab
Method: single particle / : Berndsens ZB, Rantalainen KR, Ward AB

EMDB-7859: 
Full Length HIV-1 Envelope Glycoprotein clone PC64M18C043 in complex with PGT151 Fab and PCT64-35S Fab
Method: single particle / : Berndsen ZT, Rantalainen K, Ward AB

EMDB-7860: 
HIV-1 Envelope Glycoprotein clone PC64M18C043.SOSIP.664 complex with PGT151 Fab
Method: single particle / : Berndsens ZB, Rantalainen KR, Ward AB
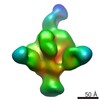
EMDB-7861: 
Full length HIV-1 Envelope Glycoprotein clone PC64M4C054 in complex with PGT151 Fab and PCT64-13C Fab
Method: single particle / : Berndsen ZT, Rantalainen K, Ward AB
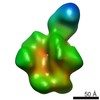
EMDB-7862: 
HIV-1 Envelope Glycoprotein clone PC64M4C054.SOSIP.664 in complex with PCT64-13F Fab
Method: single particle / : Berndsen ZT, Rantalainen K, Ward AB

EMDB-7863: 
HIV-1 Envelope Glycoprotein clone PC64M4C054.SOSIP.664 in complex with PCT64_13C Fab
Method: single particle / : Berndsens ZB, Rantalainen KR, Ward AB
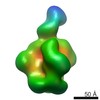
EMDB-7864: 
HIV-1 Envelope Glycoprotein clone PC64M4C054.SOSIP.664.N130A in complex with PCT64_13C Fab
Method: single particle / : Berndsen ZT, Rantalainen K, Ward AB

EMDB-7865: 
HIV-1 Envelope Glycoprotein clone PC64M4C054.SOSIP in complex with PCT64-35S Fab
Method: single particle / : Berndsens ZB, Rantalainen KR, Ward AB

EMDB-7866: 
HIV-1 Envelope Glycoprotein clone PC64M4C054.SOSIP.664.N130A in complex with PCT64_35S Fab
Method: single particle / : Berndsens ZB, Rantalainen KR, Ward AB

EMDB-8643: 
BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibodies 3BNC117 and PGT145
Method: single particle / : Lee JH, Nieusma T, Ward AB

EMDB-8644: 
BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibody 3BNC117
Method: single particle / : Lee JH, Ward AB

EMDB-2753: 
PGT124 in complex with BG505 SOSIP.664 Env trimer
Method: single particle / : Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien JP, Hua Y, Cupo A, Moore JP, Ward AB, Paulson JC, Burton DR, Wilson IA

EMDB-5918: 
Negative stain reconstruction of PGT151 Fab in complex with BG505 Env with transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB

EMDB-5919: 
Negative stain reconstruction of PGT151 Fab in complex with JR-FL Env with transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB
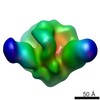
EMDB-5920: 
Negative stain reconstruction of PGT151 Fab in complex with cleaved IAVI C22 Env trimer containing the transmembrane region
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB
EMDB-5921: 
Negative stain reconstruction of PGT151 Fab in complex with the soluble Env trimer BG505 SOSIP
Method: single particle / : Lee JH, Blattner C, Wilson IA, Ward AB

EMDB-5624: 
Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans
Method: single particle / : Khayat R, Lee JH, Julien JP, Wilson IA, Ward AB
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
