-Search query
-Search result
Showing 1 - 50 of 2,085 items for (author: paul & g)

EMDB-41434: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA

EMDB-41435: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA

EMDB-41436: 
Central rod disk in D3 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L, Kirst H, Kerfeld CA

EMDB-41463: 
Synechocystis PCC 6803 Phycobilisome quenched by OCP, high resolution
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB-41475: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB-41585: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to2: 
Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8to5: 
Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8tpj: 
Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

PDB-8tro: 
Rod from high-resolution phycobilisome quenched by OCP (local refinement)
Method: single particle / : Sauer PV, Sutter M, Cupellini L

EMDB-17863: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod
Method: helical / : Hospenthal M, Zyla D, Glockshuber R, Waksman G

EMDB-17878: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod
Method: helical / : Zyla D, Hospenthal M, Glockshuber R, Waksman G

PDB-8psv: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod
Method: helical / : Hospenthal M, Zyla D, Glockshuber R, Waksman G

PDB-8ptu: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod
Method: helical / : Zyla D, Hospenthal M, Glockshuber R, Waksman G

EMDB-17375: 
Neisseria meningitidis Type IV pilus SB-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17384: 
Neisseria meningitidis Type IV pilus SB-DATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17386: 
Neisseria meningitidis Type IV pilus SA-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17683: 
Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17695: 
Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-17718: 
Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p2v: 
Neisseria meningitidis Type IV pilus SB-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p36: 
Neisseria meningitidis Type IV pilus SB-DATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8p3b: 
Neisseria meningitidis Type IV pilus SA-GATDH variant
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8pij: 
Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8piz: 
Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

PDB-8pjp: 
Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody
Method: helical / : Fernandez-Martinez D, Dumenil G

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
Method: single particle / : McCallum M, Veesler D, Seattle Structural Genomics Center for Infectious Disease (SSGCID)

EMDB-42977: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43000: 
Cryo-EM structure of SNF2h-nucleosome complex (consensus structure)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43001: 
Cryo-EM structure of SNF2h-nucleosome complex (single-bound structure)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43002: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43003: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43004: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

EMDB-43005: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

PDB-8v4y: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

PDB-8v6v: 
Cryo-EM structure of doubly-bound SNF2h-nucleosome complex
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y

PDB-8v7l: 
Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 2)
Method: single particle / : Chio US, Palovcak E, Armache JP, Narlikar GJ, Cheng Y
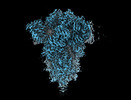
EMDB-18807: 
SD1-2 fab in complex with SARS-COV-2 BA.12.1 Spike Glycoprotein.
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI
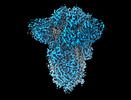
EMDB-18808: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1c: 
SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8r1d: 
SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-16903: 
60S ribosomal subunit bound to the E3-UFM1 complex (native, UFM1 pulldown)
Method: single particle / : Penchev I, DaRosa PA, Becker T, Beckmann R, Kopito R

EMDB-16680: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

PDB-8cin: 
BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI, Fry EE

EMDB-42101: 
Complex of the phosphorylated human cystic fibrosis transmembrane conductance regulator (CFTR) with CFTRinh-172 and ATP/Mg
Method: single particle / : Young PG, Fiedorczuk K, Chen J
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
