-Search query
-Search result
Showing 1 - 50 of 56 items for (author: palese & p)

EMDB-26928: 
Negative stain map of cH4/3
Method: single particle / : Han J, Ward AB
EMDB-26929: 
Negative stain half map of cH4/3 state 1
Method: single particle / : Han J, Ward AB

EMDB-26930: 
Negative stain map of cH4/3 state 3
Method: single particle / : Han J, Ward AB

EMDB-26931: 
Negative stain map of cH4/3 state 3
Method: single particle / : Han J, Ward AB

EMDB-26932: 
Negative stain map of cH4/3 state 4
Method: single particle / : Han J, Ward AB
EMDB-26933: 
Negative stain map of cH15/3
Method: single particle / : Han J, Ward AB

EMDB-26934: 
Negative stain map of cH15/3 state 1
Method: single particle / : Han J, Ward AB
EMDB-26935: 
Negative stain map of cH15/3 state 2
Method: single particle / : Han J, Ward AB

EMDB-26937: 
Negative stain map of cH15/3 state 3
Method: single particle / : Han J, Ward AB

EMDB-26938: 
Negative stain map of cH15/3 state 4
Method: single particle / : Han J, Ward AB
EMDB-26939: 
Negative stain map of cH15/3 in complex with CR9114 Fab
Method: single particle / : Han J, Ward AB

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
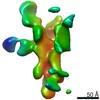
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
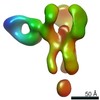
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
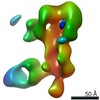
EMDB-25643: 
Negative stain map of polyclonal Fab 241.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
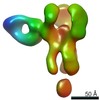
EMDB-25644: 
Negative stain map of polyclonal Fab 241.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25645: 
Negative stain map of polyclonal Fab 241.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25646: 
Negative stain map of polyclonal Fab 241.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25655: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA
Method: single particle / : Han J, Ward AB

PDB-7t3d: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA
Method: single particle / : Han J, Ward AB

EMDB-23792: 
CryoEM structure of monoclonal Fab 045-09 2B05 binding the lateral patch of influenza virus H1 HA
Method: single particle / : Han J, Ward A
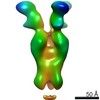
EMDB-23793: 
Negative stain map of monoclonal Fab SFV009 2G01 binding the RBS of H1 HA
Method: single particle / : Han J, Ward AB

EMDB-23794: 
Negative stain map of monoclonal Fab 045-09 2B05 binding the lateral patch of H1 HA
Method: single particle / : Han J, Ward AB

EMDB-23795: 
Negative stain map of monoclonal Fab SFV019 2A06 binding the lateral patch of H1 HA
Method: single particle / : Han J, Ward AB
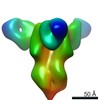
EMDB-23796: 
Negative stain map of monoclonal Fab SFV015 2F02 binding the lateral patch of H1 HA
Method: single particle / : Han J, Ward AB

EMDB-23797: 
Negative stain map of monoclonal Fab 047-09 4G02 binding the lateral patch of H1 HA
Method: single particle / : Han J, Ward AB
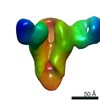
EMDB-23798: 
Negative stain map of monoclonal Fab 047-09 4B06 binding the lateral patch of H1 HA
Method: single particle / : Han J, Ward AB

PDB-7mem: 
CryoEM structure of monoclonal Fab 045-09 2B05 binding the lateral patch of influenza virus H1 HA
Method: single particle / : Han J, Ward A
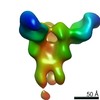
EMDB-23313: 
Negative stain map of monoclonal Fab 23 binding the lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB

EMDB-23314: 
Negative stain map of monoclonal Fab 45 binding the esterase domain of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB
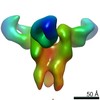
EMDB-23315: 
Negative stain map of monoclonal Fab 50 binding the lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB
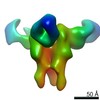
EMDB-23316: 
Negative stain map of monoclonal Fab 56 binding the lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB

EMDB-23317: 
Negative stain map of monoclonal Fab 68 binding the Sa antigenic site of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB
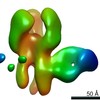
EMDB-23318: 
Negative stain map of monoclonal Fab 76 binding the stem of H1 HA
Method: single particle / : Han J, Freyn AW, Ward AB

EMDB-23319: 
CryoEM map of monoclonal Fabs 45 and 56 binding the esterase and lateral patch of H1 HA
Method: single particle / : Han J, Freyn AW, Turner HL, Ward AB

EMDB-8768: 
Negative stain of influenza B virus recombinant neuraminidase bound to 1F2 Fab
Method: single particle / : Subramaniam S, Podolsky K
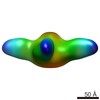
EMDB-8769: 
Negative stain of influenza B virus recombinant neuraminidase bound to 4F11 Fab
Method: single particle / : Podolsky K, Subramaniam S

EMDB-8770: 
Negative stain of influenza B virus recombinant neuraminidase
Method: single particle / : Podolsky K, Subramaniam S

EMDB-6607: 
Cryo-electron microscopy structure of influenza H1 hemagglutinin expressed on the viral surface
Method: subtomogram averaging / : Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S

EMDB-6608: 
Cryo-electron microscopy structure of chimeric influenza cH5/1 hemagglutinin expressed on the viral surface
Method: subtomogram averaging / : Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S

EMDB-6609: 
Cryo-electron microscopy structure of influenza H5 hemagglutinin expressed on the viral surface
Method: subtomogram averaging / : Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S

EMDB-6610: 
Cryo-electron microscopy structure of influenza H1 hemagglutinin bound to stalk-targeting antibody, 6F12
Method: subtomogram averaging / : Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S

EMDB-6611: 
Cryo-electron microscopy structure of chimeric influenza cH5/1 hemagglutinin bound to stalk-targeting antibody, 6F12
Method: subtomogram averaging / : Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S
EMDB-6612: 
Cryo-electron microscopy structure of influenza H1 hemagglutinin bound to head-targeting antibody, 7B2
Method: subtomogram averaging / : Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
